Get Google Scholar Profile
Arguments
- id
Your google scholar identifier. You can find it in the URL of your google scholar profile.
- sortby_date
Logical. If
TRUE, the publications are sorted by date.- use_cache
If
TRUE(default), store data to a cache file to avoid querying in next time within a day. The store file is identical for each person and each date.- cache_dir
A directory path.
Value
a Profile object with list structure.
Examples
# Put one unique Scholar ID from Google Scholar
r <- tinyscholar("FvNp0NkAAAAJ")
#> Using cache directory: /tmp/RtmpNM22mn/tinyscholar
#> Cannot find cache file /tmp/RtmpNM22mn/tinyscholar/unsorted_2022-08-09_FvNp0NkAAAAJ.rds
#> Try quering data from server: hiplot
#> Timeout/error when use hiplot server. Switch to use the server: cse.
#> Save data to cache file /tmp/RtmpNM22mn/tinyscholar/unsorted_2022-08-09_FvNp0NkAAAAJ.rds
#> Done
r
#> $publications
#> title
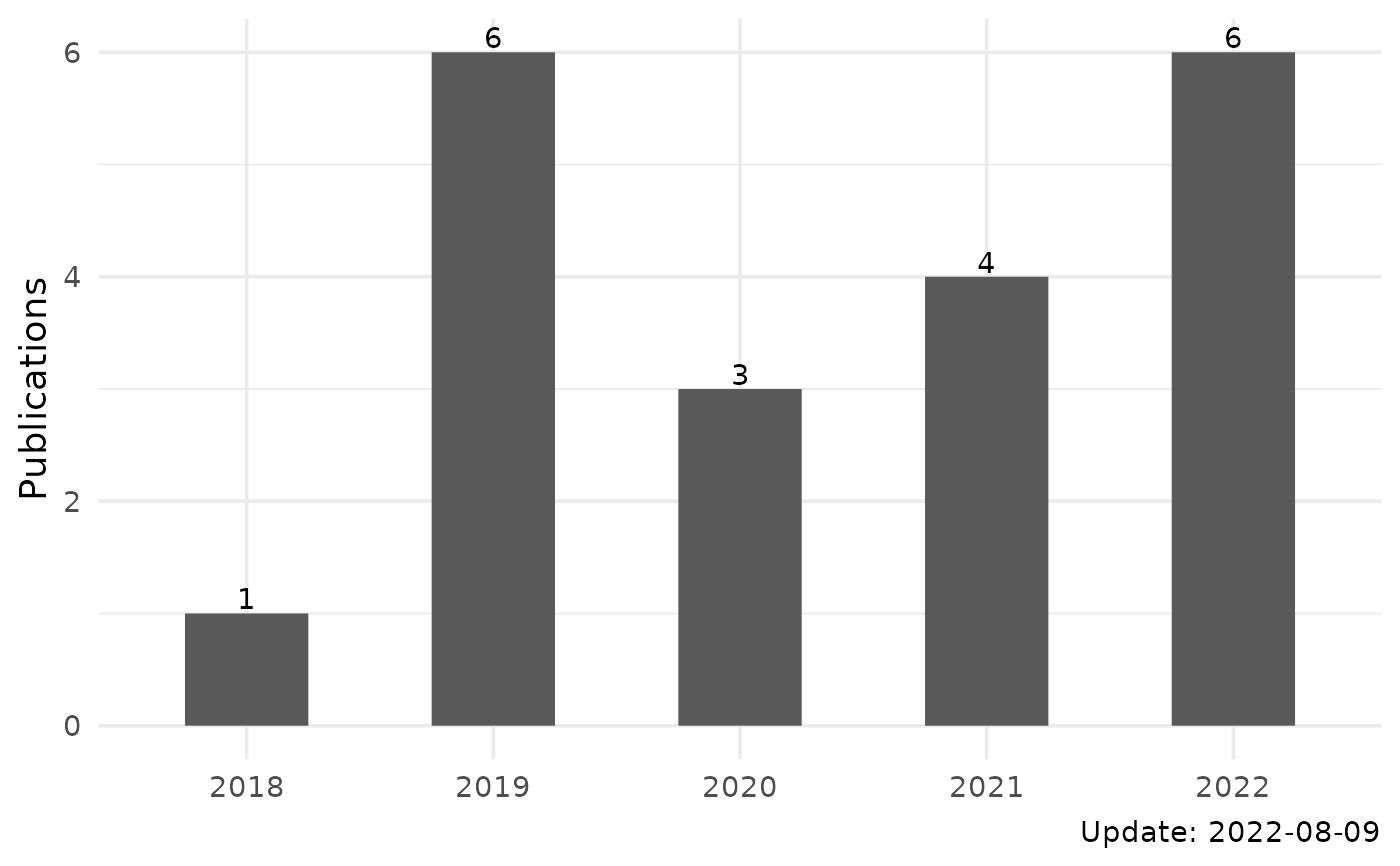
#> 1 APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer
#> 2 Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction
#> 3 Sex Differences in Cancer Immunotherapy Efficacy, Biomarkers, and Therapeutic Strategy
#> 4 The predictive power of tumor mutational burden in lung cancer immunotherapy response is influenced by patients' sex
#> 5 The UCSCXenaTools R package: a toolkit for accessing genomics data from UCSC Xena platform, from cancer multi-omics to single-cell RNA-seq
#> 6 Copy number signature analysis tool and its application in prostate cancer reveals distinct mutational processes and clinical outcomes
#> 7 UCSCXenaShiny: an R/CRAN package for interactive analysis of UCSC Xena data
#> 8 Copy number signature analyses in prostate cancer reveal distinct etiologies and clinical outcomes
#> 9 Ras downstream effector GGCT alleviates oncogenic stress
#> 10 Sigflow: an automated and comprehensive pipeline for cancer genome mutational signature analysis
#> 11 Hiplot: A comprehensive and easy-to-use web service boosting publication-ready biomedical data visualization
#> 12 Pan-cancer noncoding genomic analysis identifies functional CDC20 promoter mutation hotspots
#> 13 Can tumor mutational burden determine the most effective treatment for lung cancer patients?
#> 14 Ggct (γ‐glutamyl cyclotransferase) plays an important role in erythrocyte antioxidant defense and red blood cell survival
#> 15 Association of CSMD1 with Tumor Mutation Burden and Other Clinical Outcomes in Gastric Cancer
#> 16 Revisiting neoantigen depletion signal in the untreated cancer genome
#> 17 Deciphering clonal dynamics and metastatic routines in a rare patient of synchronous triple-primary tumors and multiple metastases with MPTevol
#> 18 Quantification of neoantigen-mediated immunoediting in cancer evolution.
#> 19 Onlinemeta: A Web Serve For Meta-Analysis Based On R-shiny
#> 20 Pan-cancer quantification of neoantigen-mediated immunoediting in cancer evolution
#> authors
#> 1 S Wang, M Jia, Z He, XS Liu
#> 2 S Wang, Z He, X Wang, H Li, XS Liu
#> 3 S Wang, LA Cowley, XS Liu
#> 4 S Wang, J Zhang, Z He, K Wu, XS Liu
#> 5 S Wang, XS Liu
#> 6 S Wang, H Li, M Song, Z Tao, T Wu, Z He, X Zhao, K Wu, XS Liu
#> 7 S Wang, Y Xiong, L Zhao, K Gu, Y Li, F Zhao, J Li, M Wang, H Wang, ...
#> 8 S Wang, H Li, M Song, Z He, T Wu, X Wang, Z Tao, K Wu, XS Liu
#> 9 Z He, S Wang, Y Shao, J Zhang, X Wu, Y Chen, J Hu, F Zhang, XS Liu
#> 10 S Wang, Z Tao, T Wu, XS Liu
#> 11 J Li, B Miao, S Wang, W Dong, H Xu, C Si, W Wang, S Duan, J Lou, Z Bao, ...
#> 12 Z He, T Wu, S Wang, J Zhang, X Sun, Z Tao, X Zhao, H Li, K Wu, XS Liu
#> 13 S Wang, Z He, X Wang, H Li, T Wu, X Sun, K Wu, XS Liu
#> 14 Z He, X Sun, S Wang, D Bai, X Zhao, Y Han, P Hao, XS Liu
#> 15 X Wang, S Wang, Y Han, M Xu, P Li, M Ke, Z Teng, P Huang, Z Diao, ...
#> 16 S Wang, X Wang, T Wu, Z He, H Li, X Sun, XS Liu
#> 17 Q Chen, QN Wu, YM Rong, S Wang, Z Zuo, L Bai, B Zhang, S Yuan, ...
#> 18 T Wu, G Wang, X Wang, S Wang, X Zhao, C Wu, W Ning, Z Tao, F Chen, ...
#> 19 Y Yi, A Lin, C Zhou, J Zhang, S Wang, P Luo
#> 20 T Wu, G Wang, X Wang, S Wang, X Zhao, C Wu, W Ning, Z Tao, F Chen, ...
#> venue citations year
#> 1 Oncogene 37 (29), 3924-3936, 2018 144 2018
#> 2 eLife, 2019 133 2019
#> 3 Molecules 24 (18), 3214, 2019 84 2019
#> 4 International journal of cancer 145 (10), 2840-2849, 2019 47 2019
#> 5 Journal of Open Source Software 4 (40), 1627, 2019 29 2019
#> 6 PLoS genetics 17 (5), e1009557, 2021 21 2021
#> 7 Bioinformatics 38 (2), 527-529, 2022 18 2022
#> 8 medRxiv, 2020 8 2020
#> 9 Iscience 19, 256-266, 2019 8 2019
#> 10 Bioinformatics 37 (11), 1590-1592, 2020 6 2020
#> 11 bioRxiv, 2022 4 2022
#> 12 Iscience 24 (4), 102285, 2021 2 2021
#> 13 Lung Cancer Management 8 (4), LMT21, 2019 2 2019
#> 14 British Journal of Haematology 195 (2), 267-275, 2021 1 2021
#> 15 International Journal of General Medicine 14, 8293, 2021 1 2021
#> 16 bioRxiv, 2020 1 2020
#> 17 Briefings in Bioinformatics, 2022 0 2022
#> 18 Cancer Research, 2022 0 2022
#> 19 bioRxiv, 2022 0 2022
#> 20 bioRxiv, 2022 0 2022
#>
#> $citations
#> when count
#> 1 total 509
#> 2 2018 2
#> 3 2019 36
#> 4 2020 117
#> 5 2021 195
#> 6 2022 154
#>
#> attr(,"class")
#> [1] "ScholarProfile" "list"
if (!is.null(r)) {
tb <- scholar_table(r)
tb$citations
tb$publications
pl <- scholar_plot(r)
pl$citations
pl$publications
}