Plot Reference (Mainly COSMIC) Signature Profile
Source:R/show_cosmic_signature_profile.R
show_cosmic_sig_profile.RdPlot Reference (Mainly COSMIC) Signature Profile
show_cosmic_sig_profile(
sig_index = NULL,
show_index = TRUE,
sig_db = "legacy",
...
)Arguments
- sig_index
a vector for signature index. "ALL" for all signatures.
- show_index
if
TRUE, show valid indices.- sig_db
default 'legacy', it can be 'legacy' (for COSMIC v2 'SBS'), 'SBS', 'DBS', 'ID' and 'TSB' (for COSMIV v3.1 signatures) for small scale mutations. For more specific details, it can also be 'SBS_hg19', 'SBS_hg38', 'SBS_mm9', 'SBS_mm10', 'DBS_hg19', 'DBS_hg38', 'DBS_mm9', 'DBS_mm10' to use COSMIC v3 reference signatures from Alexandrov, Ludmil B., et al. (2020) (reference #1). In addition, it can be one of "SBS_Nik_lab_Organ", "RS_Nik_lab_Organ", "SBS_Nik_lab", "RS_Nik_lab" to refer reference signatures from Degasperi, Andrea, et al. (2020) (reference #2); "RS_BRCA560", "RS_USARC" to reference signatures from BRCA560 and USARC cohorts; "CNS_USARC" (40 categories), "CNS_TCGA" (48 categories) to reference copy number signatures from USARC cohort and TCGA; "CNS_TCGA176" (176 categories) and "CNS_PCAWG176" (176 categories) to reference copy number signatures from PCAWG and TCGA separately. UPDATE, the latest version of reference version can be automatically downloaded and loaded from https://cancer.sanger.ac.uk/signatures/downloads/ when a option with
latest_prefix is specified (e.g. "latest_SBS_GRCh37"). Note: the signature profile for different genome builds are basically same. And specific database (e.g. 'SBS_mm10') contains less signatures than all COSMIC signatures (because some signatures are not detected from Alexandrov, Ludmil B., et al. (2020)). For all available options, check the parameter setting.- ...
other arguments passing to show_sig_profile.
Value
a ggplot object
Examples
# \donttest{
show_cosmic_sig_profile()
#>
#> Valid index for db 'legacy':
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30
show_cosmic_sig_profile(sig_db = "SBS")
#>
#> Valid index for db 'SBS':
#> 1 2 3 4 5 6 7a 7b 7c 7d 8 9 10a 10b 11 12 13 14 15 16 17a 17b 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 84 85 86 87 88 89 90
show_cosmic_sig_profile(sig_index = 1:5)
#>
#> Valid index for db 'legacy':
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30
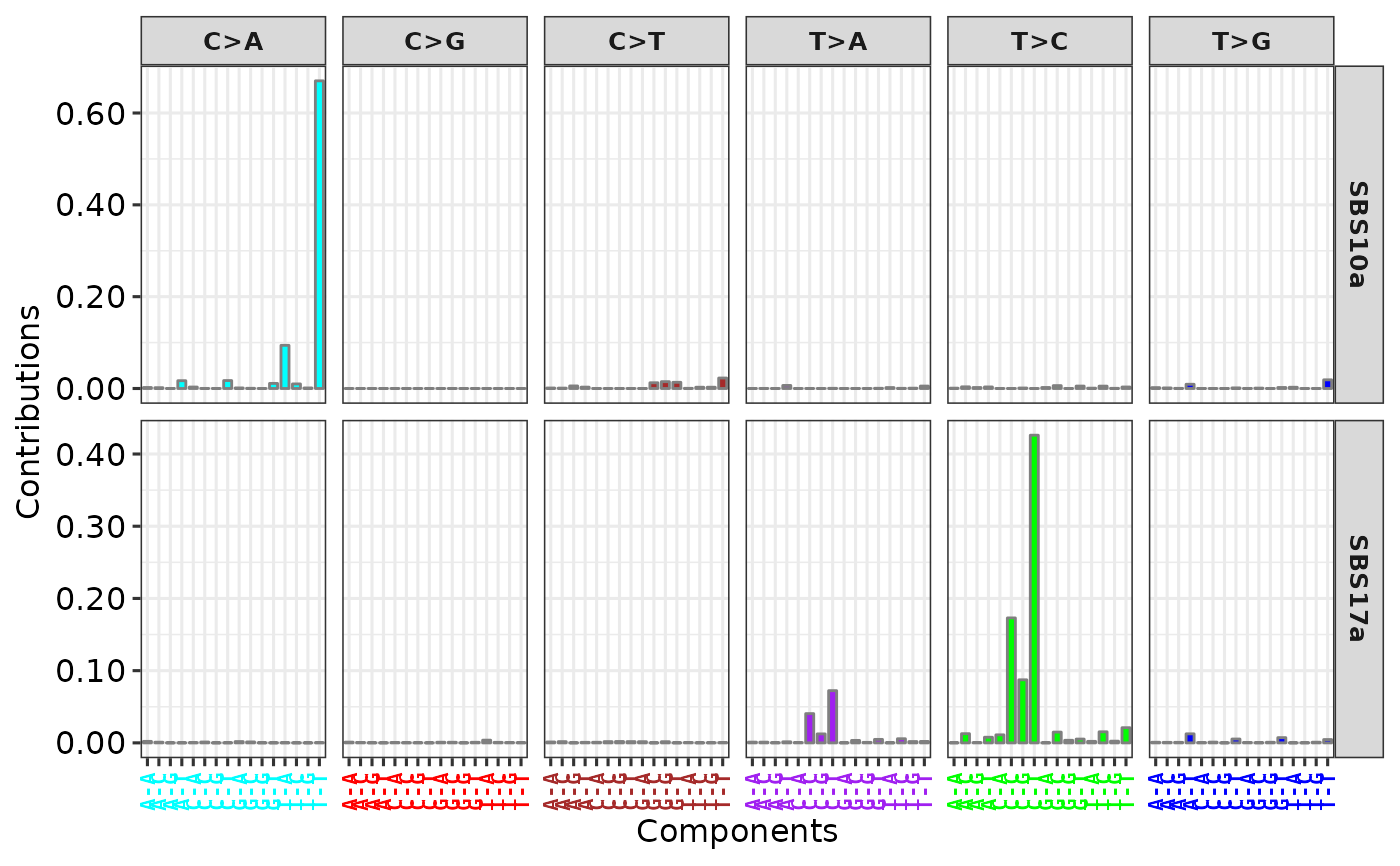
 show_cosmic_sig_profile(sig_db = "SBS", sig_index = c("10a", "17a"))
#>
#> Valid index for db 'SBS':
#> 1 2 3 4 5 6 7a 7b 7c 7d 8 9 10a 10b 11 12 13 14 15 16 17a 17b 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 84 85 86 87 88 89 90
show_cosmic_sig_profile(sig_db = "SBS", sig_index = c("10a", "17a"))
#>
#> Valid index for db 'SBS':
#> 1 2 3 4 5 6 7a 7b 7c 7d 8 9 10a 10b 11 12 13 14 15 16 17a 17b 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 84 85 86 87 88 89 90
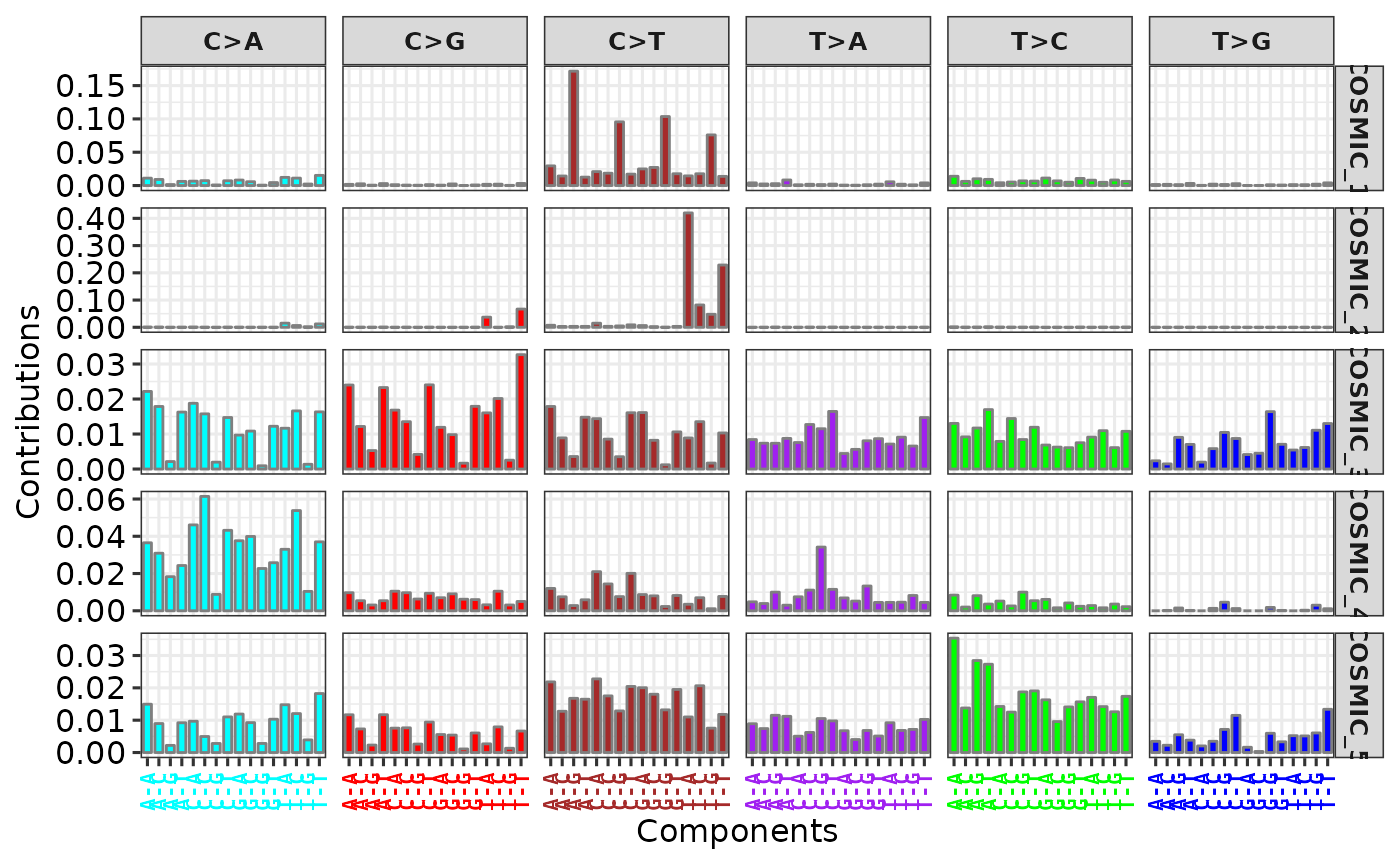
 gg <- show_cosmic_sig_profile(sig_index = 1:5)
#>
#> Valid index for db 'legacy':
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30
gg$aetiology
#> aetiology
#> COSMIC_1 spontaneous deamination of 5-methylcytosine
#> COSMIC_2 APOBEC Cytidine Deaminase (C>T)
#> COSMIC_3 defects in DNA-DSB repair by HR
#> COSMIC_4 exposure to tobacco (smoking) mutagens
#> COSMIC_5 Unknown
# }
gg <- show_cosmic_sig_profile(sig_index = 1:5)
#>
#> Valid index for db 'legacy':
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30
gg$aetiology
#> aetiology
#> COSMIC_1 spontaneous deamination of 5-methylcytosine
#> COSMIC_2 APOBEC Cytidine Deaminase (C>T)
#> COSMIC_3 defects in DNA-DSB repair by HR
#> COSMIC_4 exposure to tobacco (smoking) mutagens
#> COSMIC_5 Unknown
# }