Show Cox Models
show_models(
models,
model_names = NULL,
covariates = NULL,
merge_models = FALSE,
drop_controls = FALSE,
headings = list(variable = "Variable", n = "N", measure = "Hazard ratio", ci = NULL,
p = "p"),
...
)Arguments
- models
a
ezcox_modelsfromget_models()or a (named) list of Cox models.- model_names
model names to show when
merge_models=TRUE.- covariates
a character vector optionally listing the variables to include in the plot (defaults to all variables).
- merge_models
if 'TRUE', merge all models and keep the plot tight.
- drop_controls
works when
covariates=NULLandmodelsis aezcox_models, ifTRUE, it removes control variables automatically.- headings
a
listfor setting the heading text.- ...
other arguments passing to
forestmodel::forest_model().
Value
a ggplot object
Examples
library(survival)
zz <- ezcox(lung, covariates = c("sex", "ph.ecog"), controls = "age", return_models = TRUE)
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
mds <- get_models(zz)
show_models(mds)
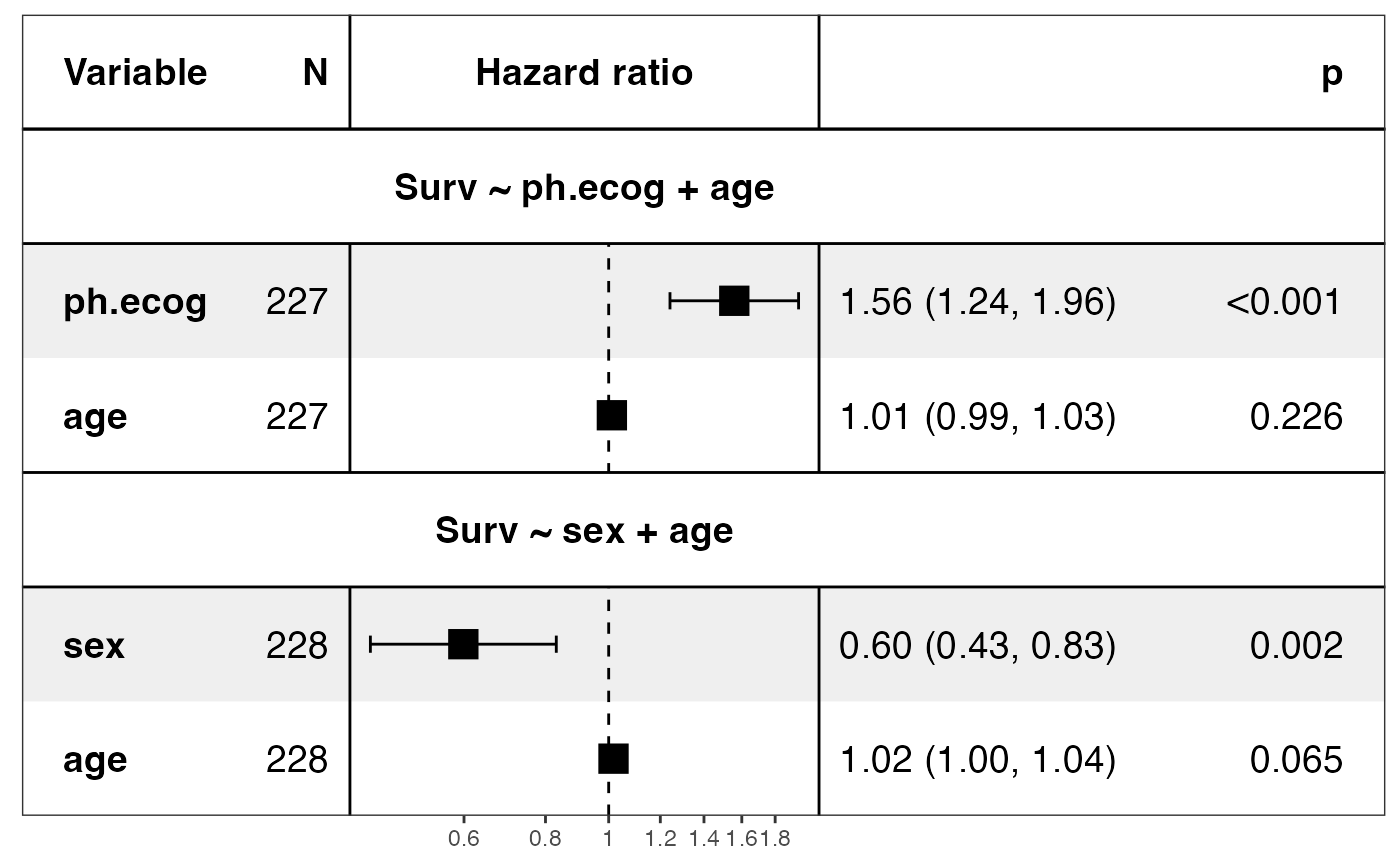 show_models(mds, model_names = paste0("Model ", 1:2))
show_models(mds, model_names = paste0("Model ", 1:2))
 show_models(mds, covariates = c("sex", "ph.ecog"))
show_models(mds, covariates = c("sex", "ph.ecog"))
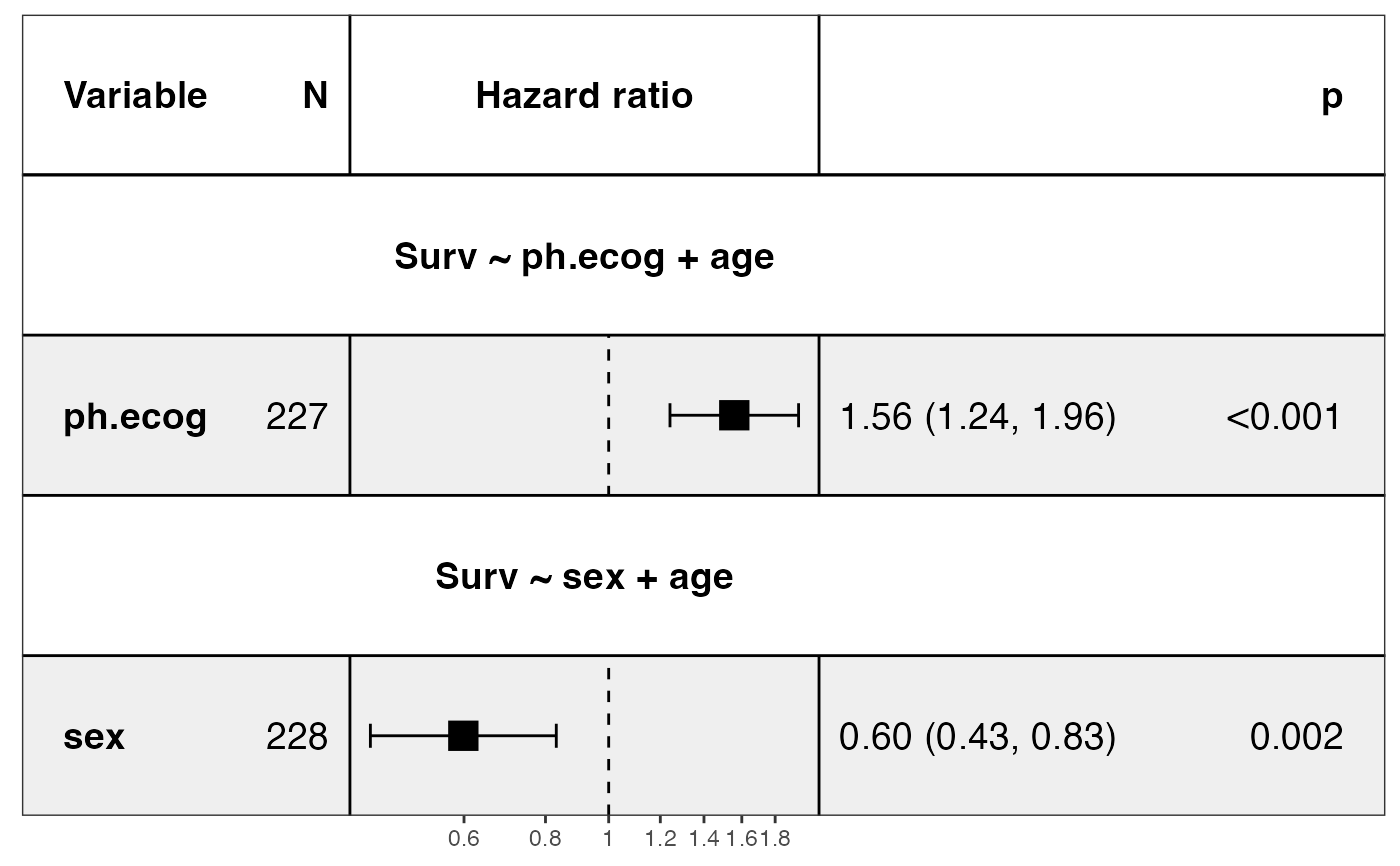 show_models(mds, drop_controls = TRUE)
#> covariates=NULL but drop_controls=TRUE, detecting controls...
#> Yes. Setting variables to keep...
#> Done.
show_models(mds, drop_controls = TRUE)
#> covariates=NULL but drop_controls=TRUE, detecting controls...
#> Yes. Setting variables to keep...
#> Done.
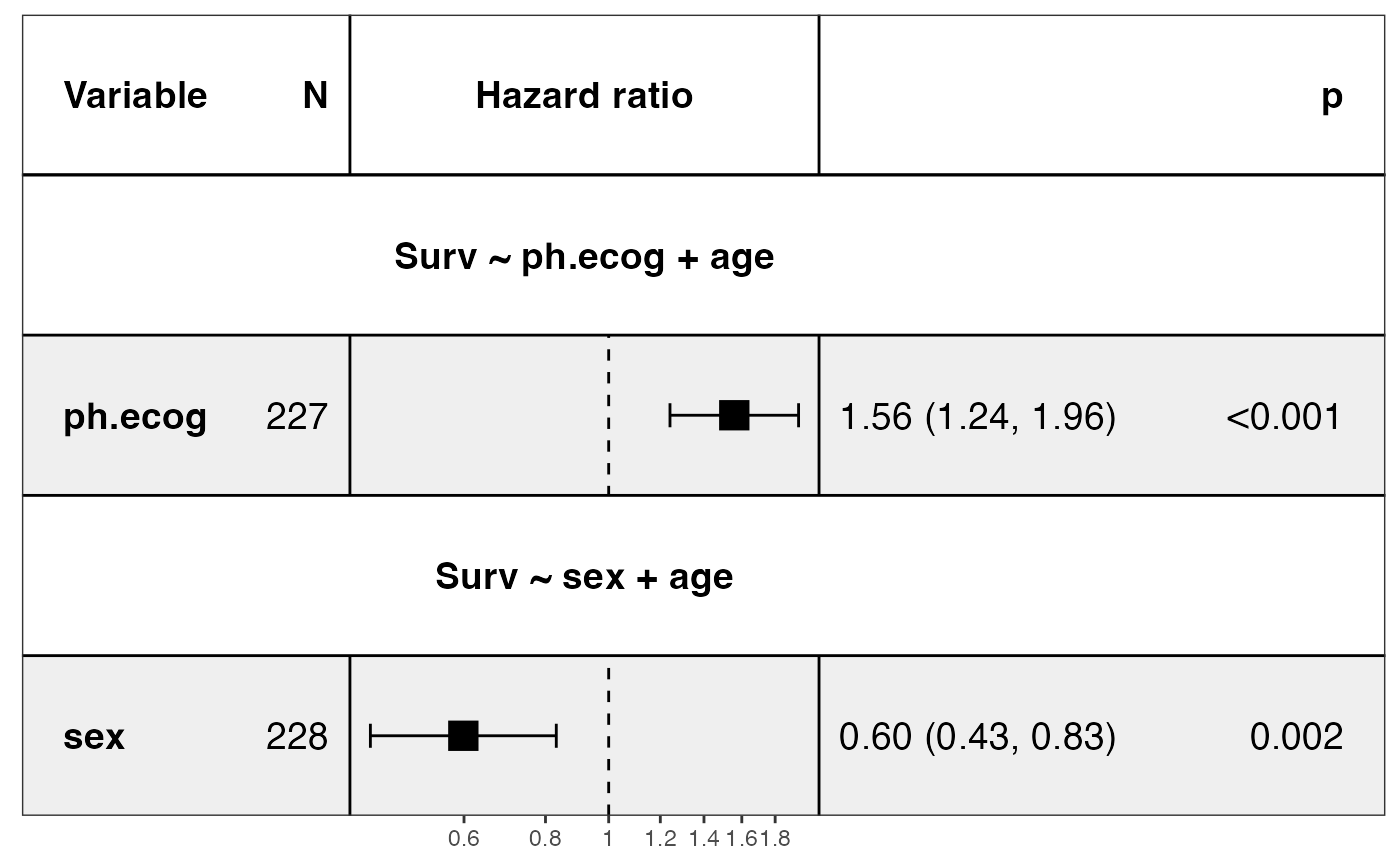 show_models(mds, merge_models = TRUE)
show_models(mds, merge_models = TRUE)
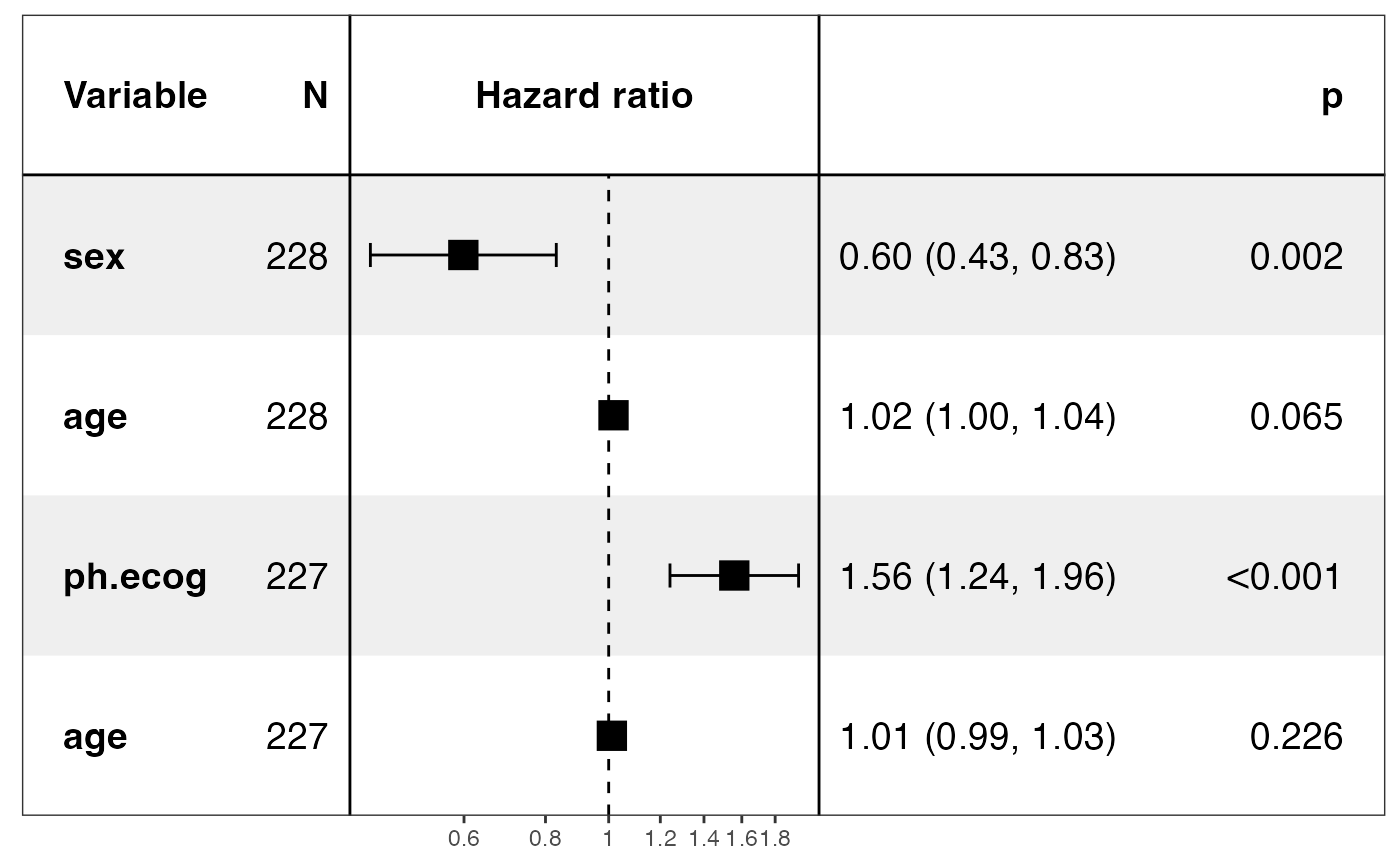 p <- show_models(mds, merge_models = TRUE, drop_controls = TRUE)
#> covariates=NULL but drop_controls=TRUE, detecting controls...
#> Yes. Setting variables to keep...
#> Done.
p
p <- show_models(mds, merge_models = TRUE, drop_controls = TRUE)
#> covariates=NULL but drop_controls=TRUE, detecting controls...
#> Yes. Setting variables to keep...
#> Done.
p
