ezcox: Easily Show Cox Forestplot in One Command
Shixiang Wang SYSUCC
2023-05-08
Source:vignettes/ezforest.Rmd
ezforest.Rmd
library(survival)
library(ezcox)
#> Welcome to 'ezcox' package!
#> =======================================================================
#> You are using ezcox version 1.0.4
#>
#> Project home : https://github.com/ShixiangWang/ezcox
#> Documentation: https://shixiangwang.github.io/ezcox
#> Cite as : arXiv:2110.14232
#> =======================================================================
#> forester
For simple and general forest data, you can use
forester(), it is lightweight and can be applied to any
proper data (not limited to Cox model).
t1 <- ezcox(lung, covariates = c(
"age", "sex",
"ph.karno", "pat.karno"
))
#> => Processing variable age
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.karno
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable pat.karno
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
p <- forester(t1, xlim = c(0, 1.5))
p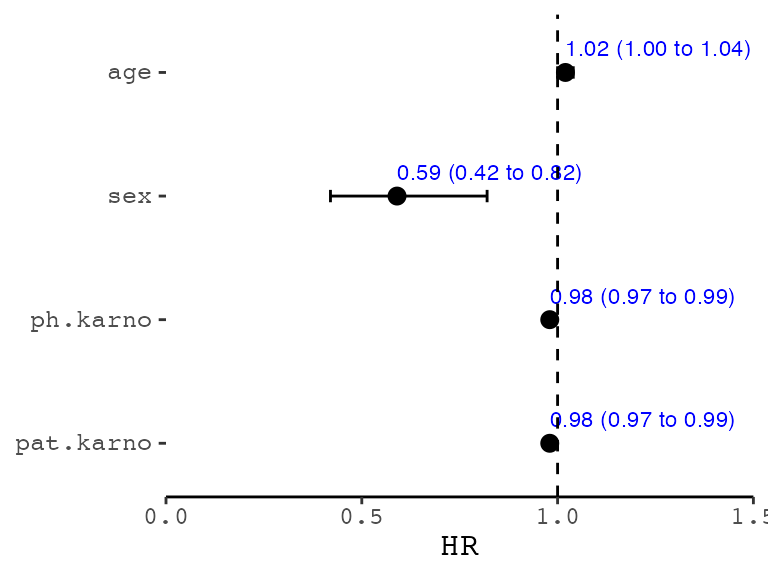
show_forest
For more powerful plot features, you need to use
show_forest(). Unlike the forester(), the
ezcox() is included in the function.
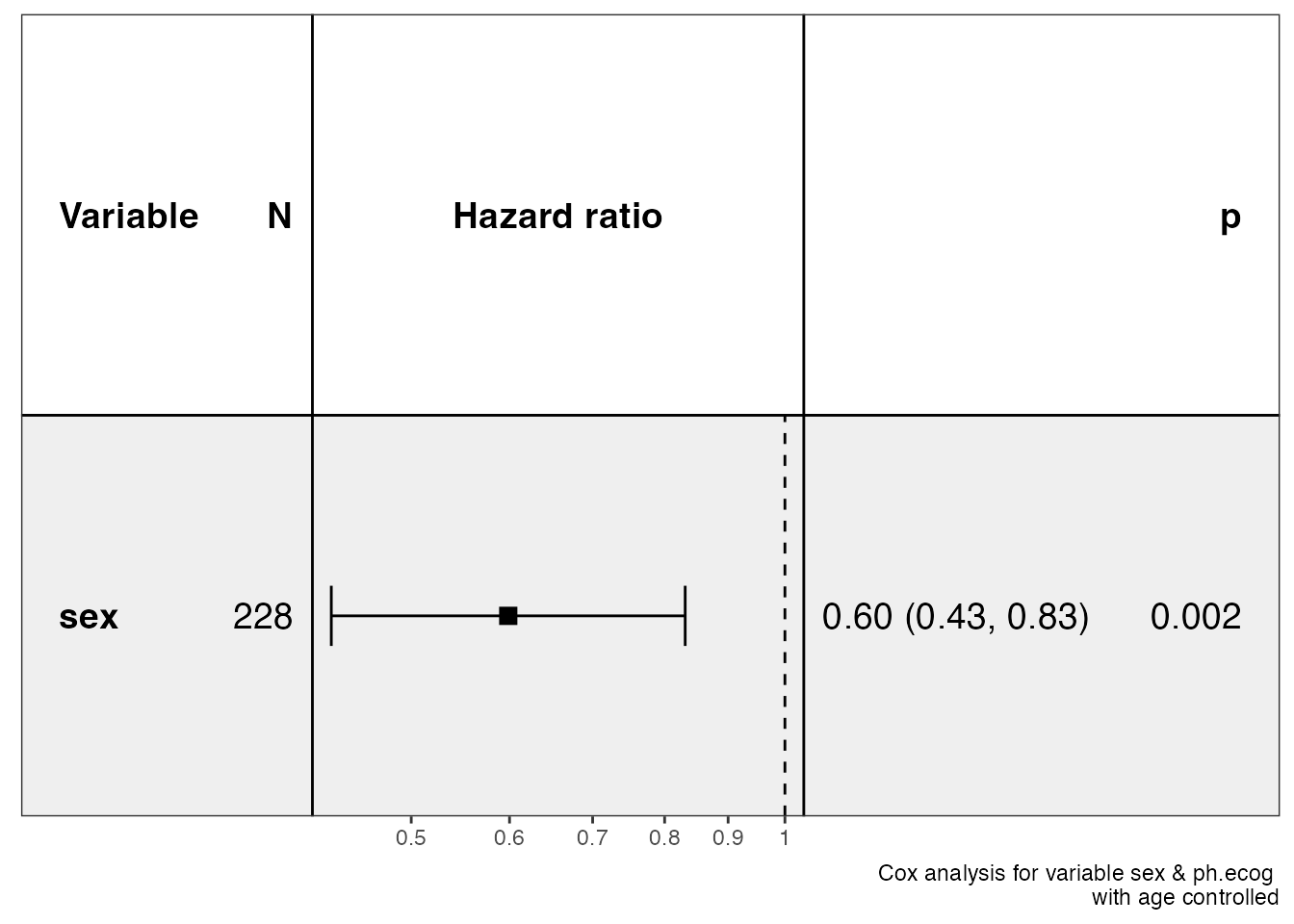
show_forest(lung, covariates = c("sex", "ph.ecog"), controls = "age")
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.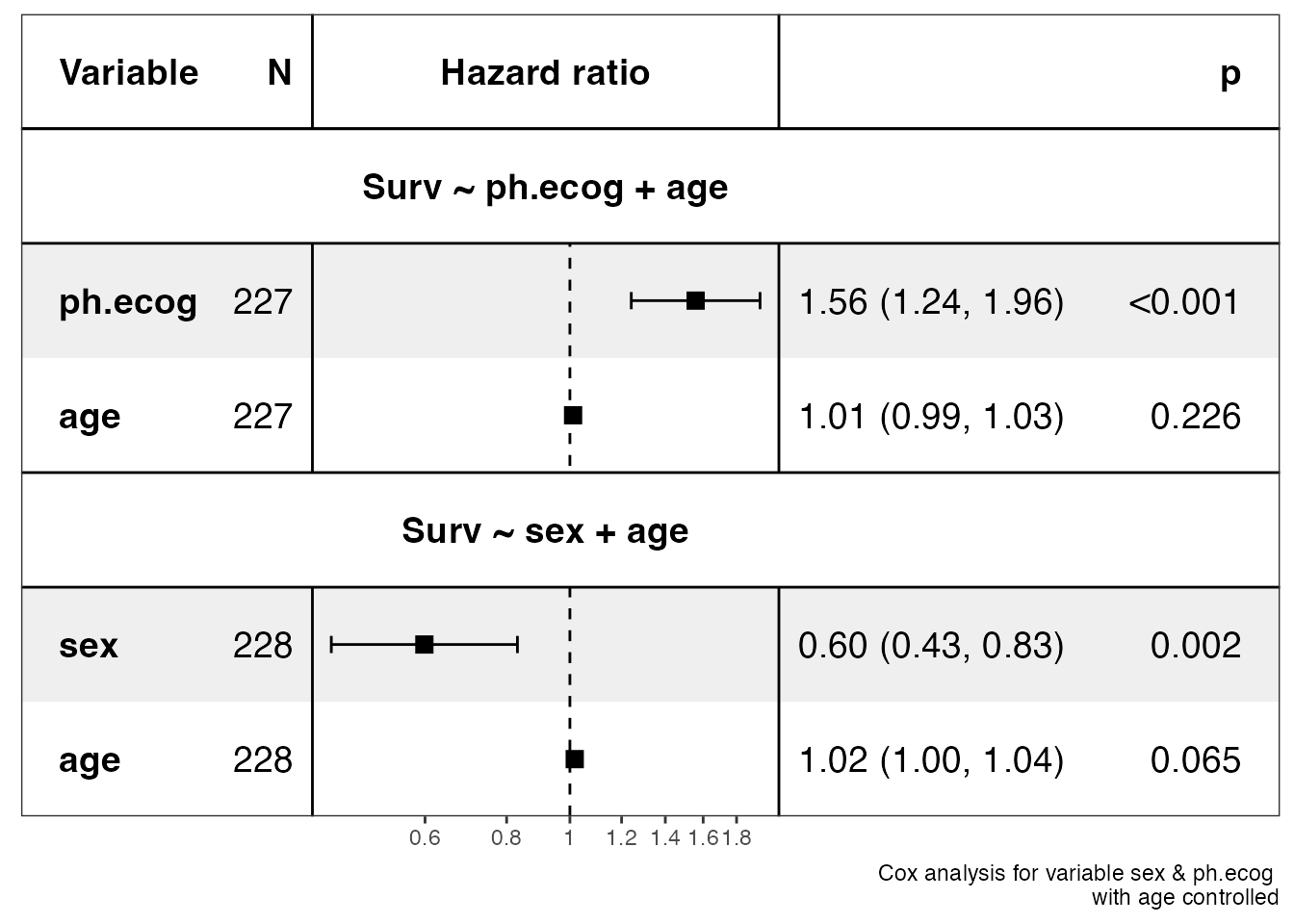
show_forest(lung, covariates = c("sex", "ph.ecog"), controls = "age", merge_models = TRUE)
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.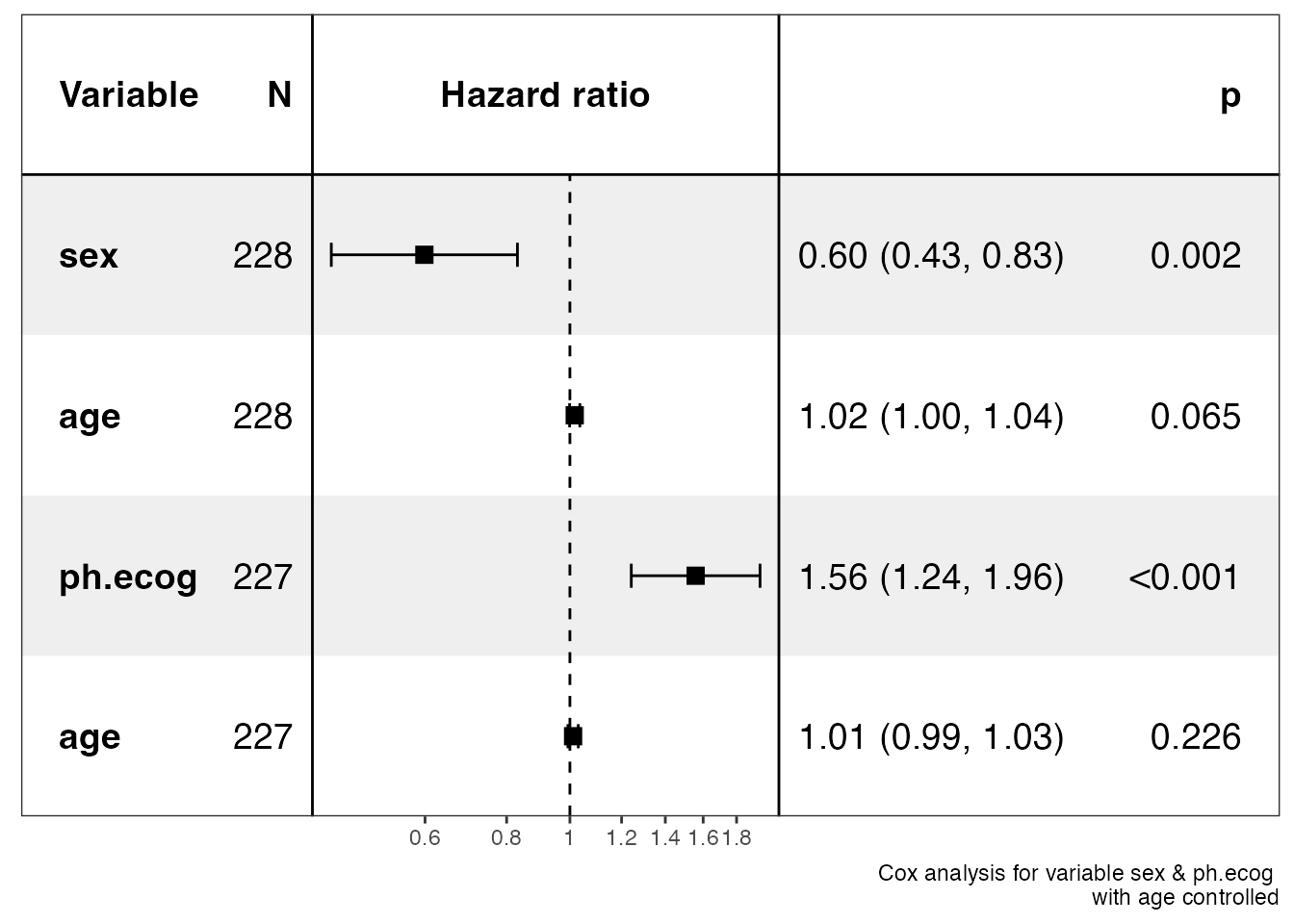
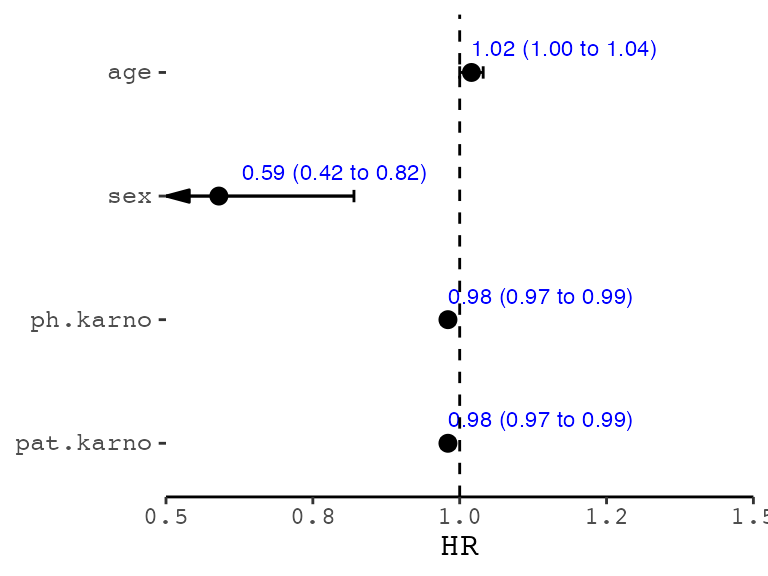
show_forest(lung,
covariates = c("sex", "ph.ecog"), controls = "age", merge_models = TRUE,
drop_controls = TRUE
)
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> covariates=NULL but drop_controls=TRUE, detecting controls...
#> Yes. Setting variables to keep...
#> Done.
show_forest(lung,
covariates = c("sex", "ph.ecog"), controls = "age", merge_models = TRUE,
vars_to_show = "sex"
)
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> Resized limits to included dashed line in forest panel