This is a wrapper of function ezcox, get_models and show_models. It focus on generating forest plot easily and flexibly.
show_forest(
data,
covariates,
controls = NULL,
time = "time",
status = "status",
merge_models = FALSE,
model_names = NULL,
vars_to_show = NULL,
drop_controls = FALSE,
add_caption = TRUE,
point_size = 3,
point_shape = 15,
color = "red",
banded = TRUE,
headings = list(variable = "Variable", n = "N", measure = "Hazard ratio", ci = NULL,
p = "p"),
model_dir = file.path(tempdir(), "ezcox"),
verbose = TRUE,
...
)Arguments
- data
a
data.framecontaining variables, time and os status.- covariates
a character vector optionally listing the variables to include in the plot (defaults to all variables).
- controls
column names specifying controls. The names with pattern "*:|()" will be treated as interaction/combination term, please make sure all column names in
dataare valid R variable names.- time
column name specifying time, default is 'time'.
- status
column name specifying event status, default is 'status'.
- merge_models
if 'TRUE', merge all models and keep the plot tight.
- model_names
model names to show when
merge_models=TRUE.- vars_to_show
default is
NULL, show all variables (including controls). You can use this to choose variables to show, but remember, the models have not been changed.- drop_controls
works when
covariates=NULLandmodelsis aezcox_models, ifTRUE, it removes control variables automatically.- add_caption
if
TRUE, add caption to the plot.- point_size
size of point.
- point_shape
shape value of point.
- color
color for point and segment.
- banded
if
TRUE(default), create banded background color.- headings
a
listfor setting the heading text.- model_dir
a path for storing model results.
- verbose
if
TRUE, print extra info.- ...
other arguments passing to
forestmodel::forest_model().
Value
a ggplot object
Examples
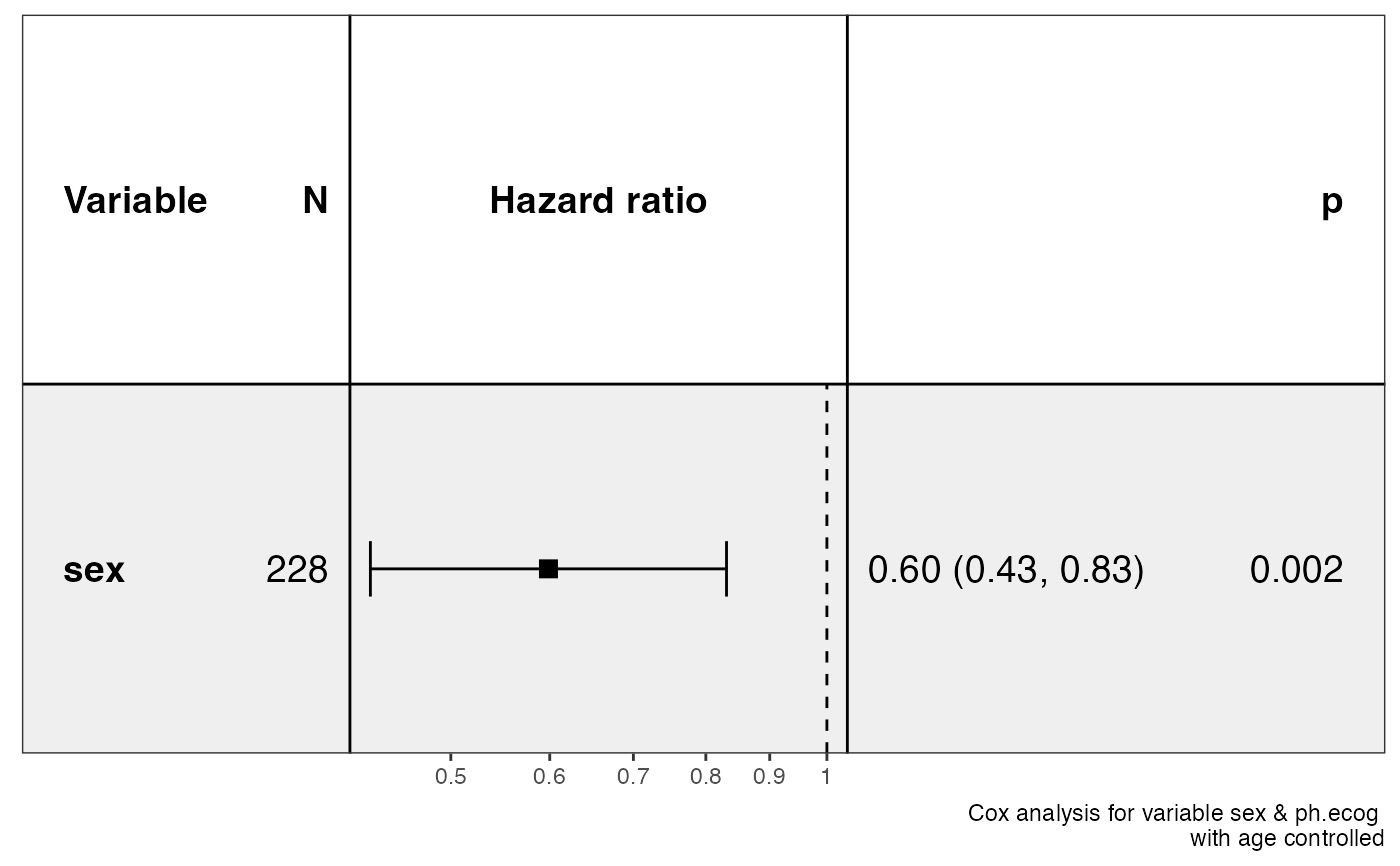
library(survival)
show_forest(lung, covariates = c("sex", "ph.ecog"), controls = "age")
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
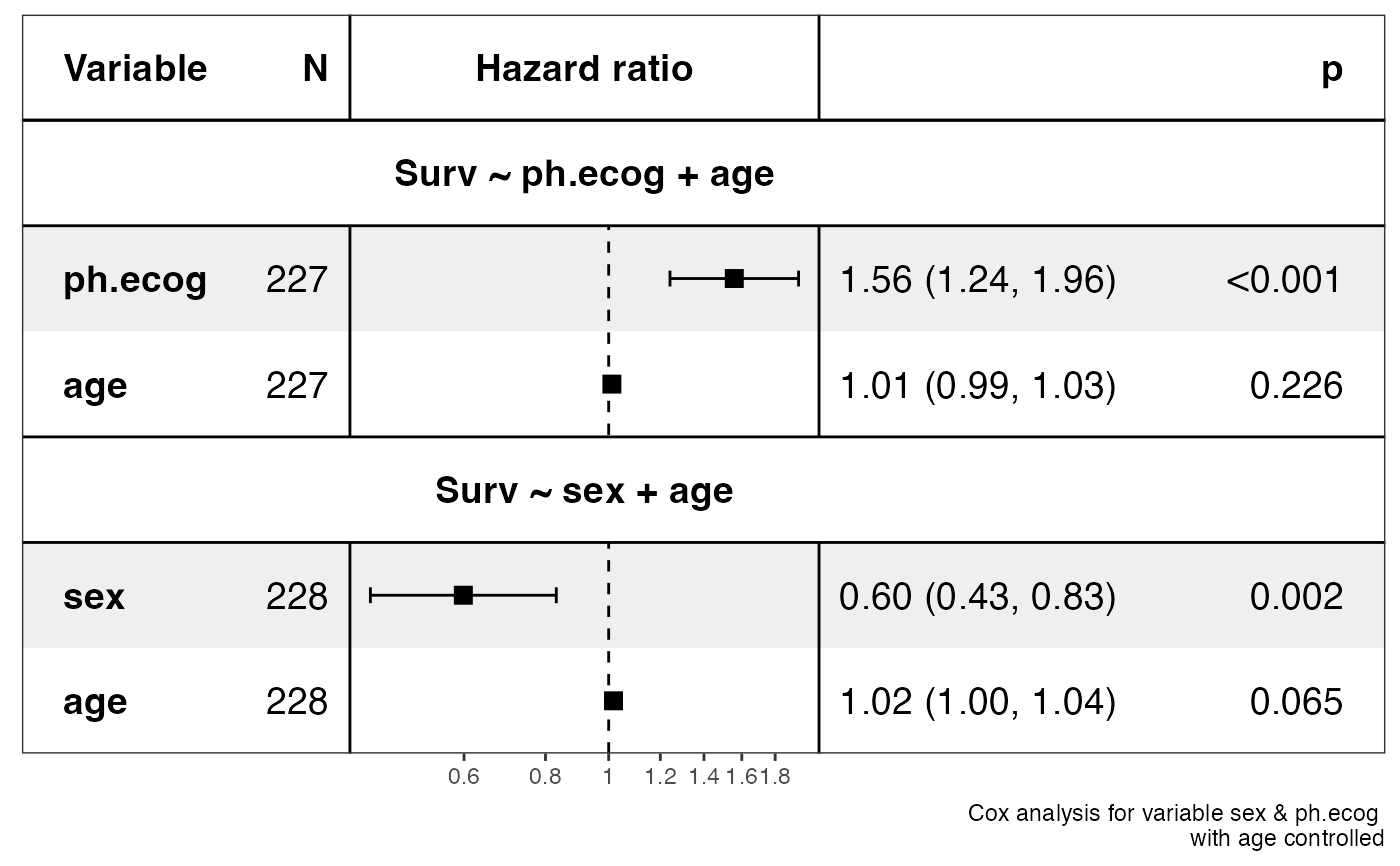 show_forest(lung, covariates = c("sex", "ph.ecog"), controls = "age", merge_models = TRUE)
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
show_forest(lung, covariates = c("sex", "ph.ecog"), controls = "age", merge_models = TRUE)
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
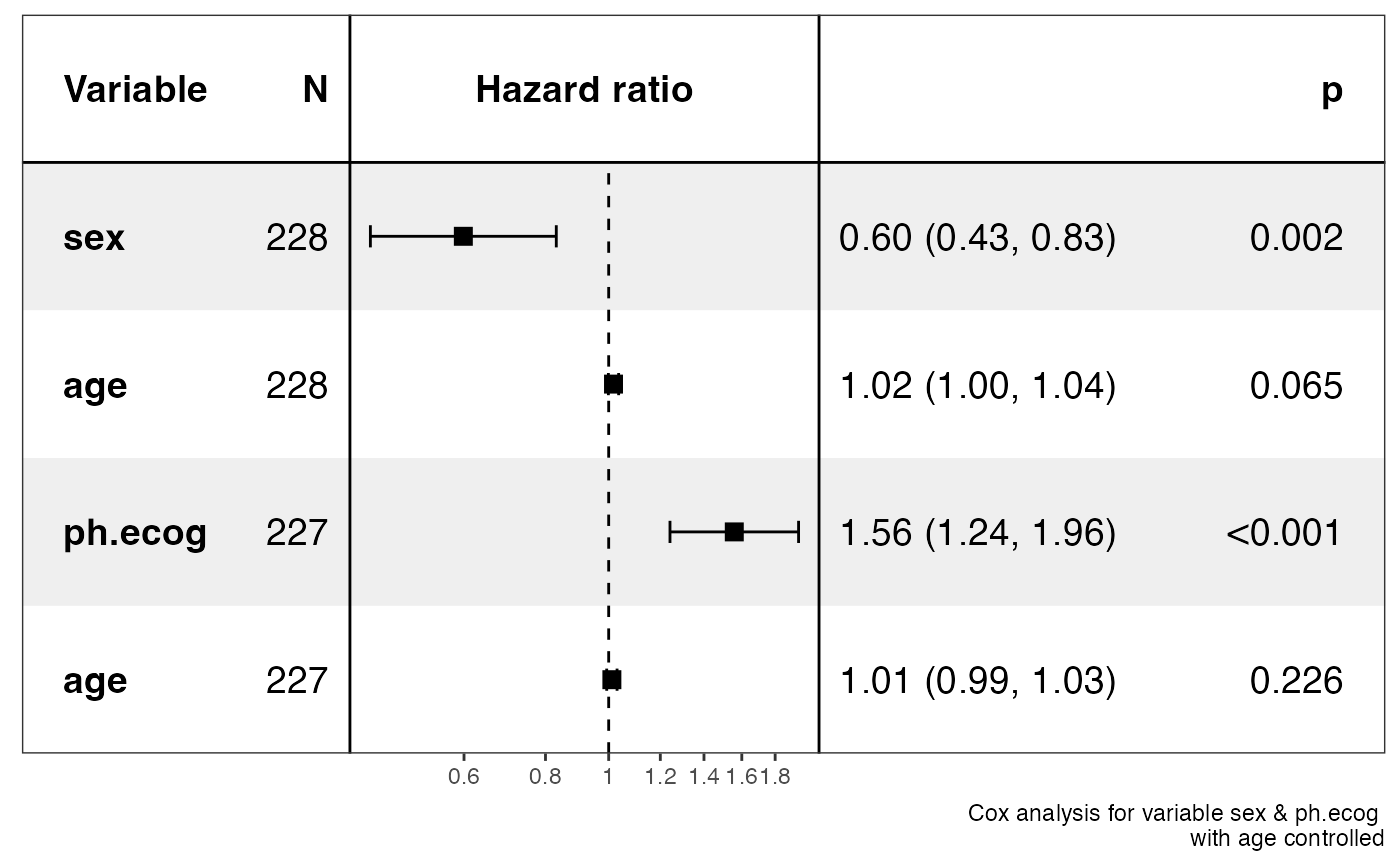 show_forest(lung,
covariates = c("sex", "ph.ecog"), controls = "age", merge_models = TRUE,
drop_controls = TRUE
)
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> covariates=NULL but drop_controls=TRUE, detecting controls...
#> Yes. Setting variables to keep...
#> Done.
show_forest(lung,
covariates = c("sex", "ph.ecog"), controls = "age", merge_models = TRUE,
drop_controls = TRUE
)
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> covariates=NULL but drop_controls=TRUE, detecting controls...
#> Yes. Setting variables to keep...
#> Done.
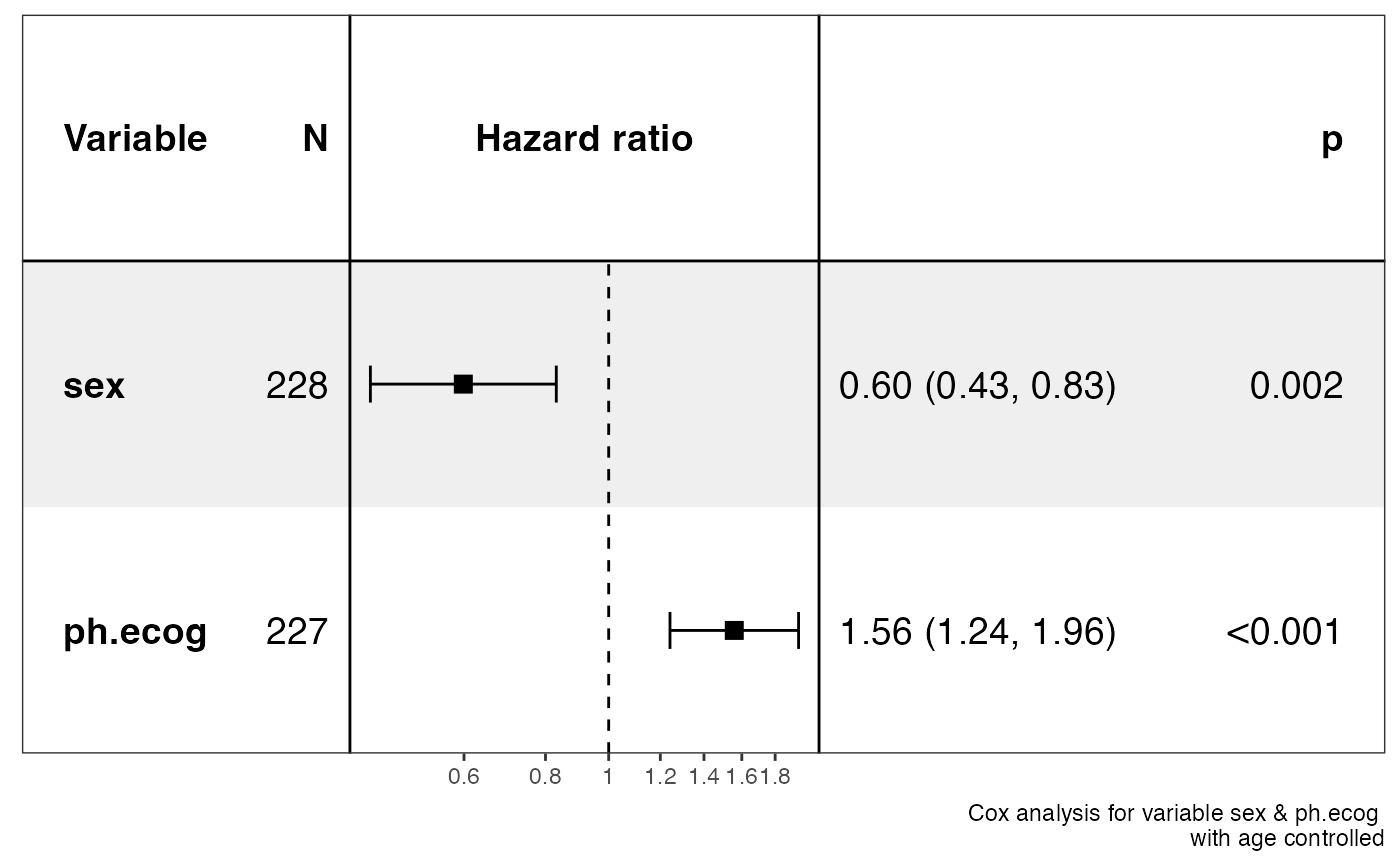 p <- show_forest(lung,
covariates = c("sex", "ph.ecog"), controls = "age", merge_models = TRUE,
vars_to_show = "sex"
)
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> Resized limits to included dashed line in forest panel
p
p <- show_forest(lung,
covariates = c("sex", "ph.ecog"), controls = "age", merge_models = TRUE,
vars_to_show = "sex"
)
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> Resized limits to included dashed line in forest panel
p