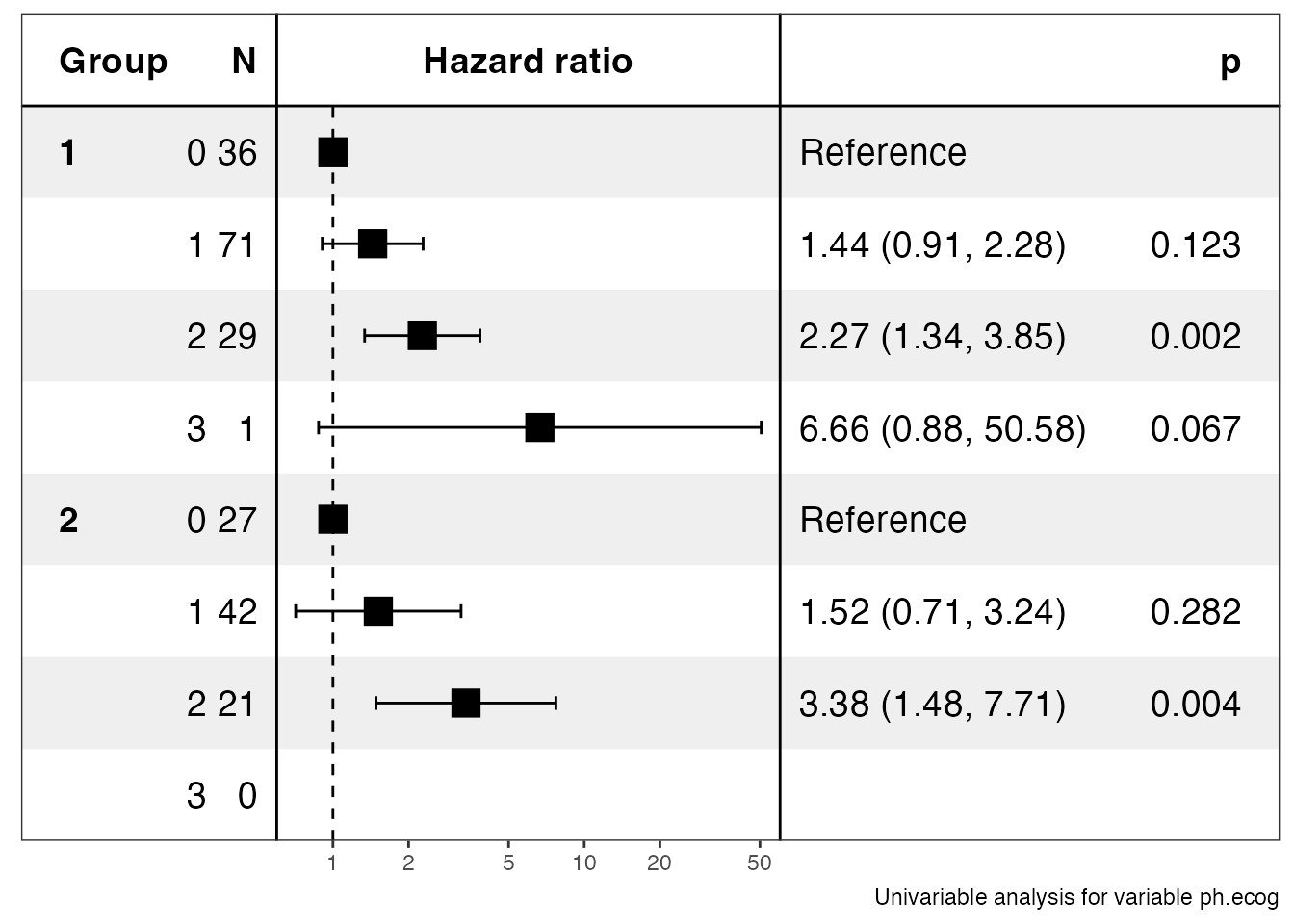
library(ezcox)
#> Welcome to 'ezcox' package!
#> =======================================================================
#> You are using ezcox version 1.0.4
#>
#> Project home : https://github.com/ShixiangWang/ezcox
#> Documentation: https://shixiangwang.github.io/ezcox
#> Cite as : arXiv:2110.14232
#> =======================================================================
#>
library(survival)
lung$ph.ecog <- factor(lung$ph.ecog)
ezcox_group(lung, grp_var = "sex", covariate = "ph.ecog")
#> => Processing variable 1
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable 2
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> covariates=NULL but drop_controls=TRUE, detecting controls...
#> No. Skipping...
#> Done.
#> Returns a list containing data and ggplot.
#> $data
#> $stats
#> # A tibble: 6 × 13
#> Group Variable is_control contras…¹ ref_l…² n_con…³ n_ref beta HR lower…⁴
#> <chr> <chr> <lgl> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 ph.ecog FALSE 1 0 71 36 0.364 1.44 0.907
#> 2 1 ph.ecog FALSE 2 0 29 36 0.819 2.27 1.34
#> 3 1 ph.ecog FALSE 3 0 1 36 1.9 6.66 0.877
#> 4 2 ph.ecog FALSE 1 0 42 27 0.416 1.52 0.711
#> 5 2 ph.ecog FALSE 2 0 21 27 1.22 3.38 1.48
#> 6 2 ph.ecog FALSE 3 0 0 27 NA NA NA
#> # … with 3 more variables: upper_95 <dbl>, p.value <dbl>, global.pval <dbl>,
#> # and abbreviated variable names ¹contrast_level, ²ref_level, ³n_contrast,
#> # ⁴lower_95
#>
#> $models
#> # A tibble: 2 × 6
#> Group Variable control model_file model status
#> <chr> <chr> <chr> <chr> <list> <lgl>
#> 1 1 ph.ecog NA /var/folders/bj/nw1w4g1j37ddpgb6zmh3sfh… <coxph> TRUE
#> 2 2 ph.ecog NA /var/folders/bj/nw1w4g1j37ddpgb6zmh3sfh… <coxph> TRUE
#>
#> attr(,"class")
#> [1] "ezcox"
#>
#> $plot
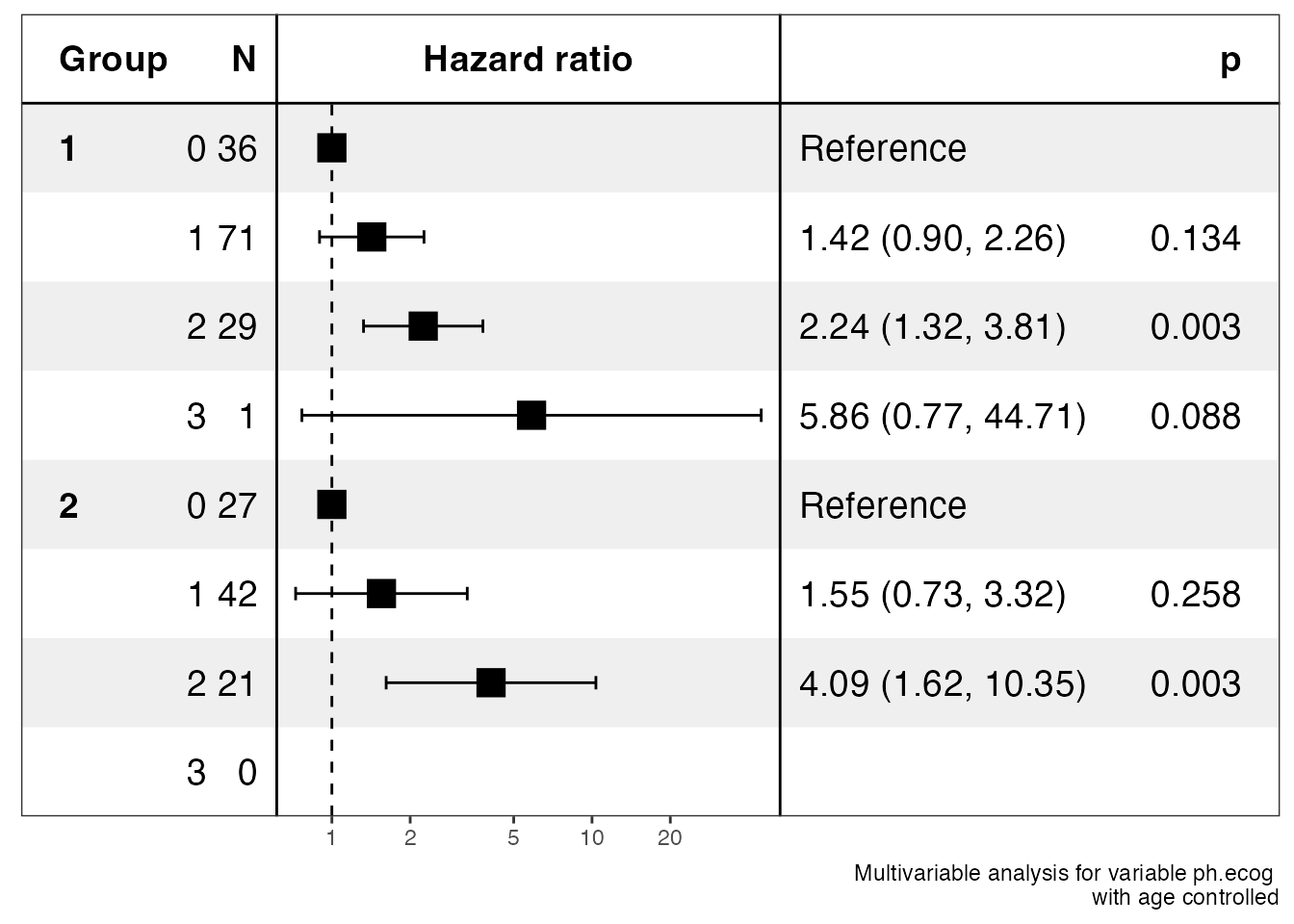
ezcox_group(lung, grp_var = "sex", covariate = "ph.ecog", controls = "age")
#> => Processing variable 1
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable 2
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> covariates=NULL but drop_controls=TRUE, detecting controls...
#> Yes. Setting variables to keep...
#> Done.
#> Returns a list containing data and ggplot.
#> $data
#> $stats
#> # A tibble: 8 × 13
#> Group Variable is_control contr…¹ ref_l…² n_con…³ n_ref beta HR lower…⁴
#> <chr> <chr> <lgl> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 ph.ecog FALSE 1 0 71 36 0.353 1.42 0.897
#> 2 1 ph.ecog FALSE 2 0 29 36 0.809 2.24 1.32
#> 3 1 ph.ecog FALSE 3 0 1 36 1.77 5.86 0.767
#> 4 1 ph.ecog TRUE age age 138 138 0.0182 1.02 0.996
#> 5 2 ph.ecog FALSE 1 0 42 27 0.439 1.55 0.726
#> 6 2 ph.ecog FALSE 2 0 21 27 1.41 4.09 1.62
#> 7 2 ph.ecog FALSE 3 0 0 27 NA NA NA
#> 8 2 ph.ecog TRUE age age 90 90 -0.0153 0.985 0.952
#> # … with 3 more variables: upper_95 <dbl>, p.value <dbl>, global.pval <dbl>,
#> # and abbreviated variable names ¹contrast_level, ²ref_level, ³n_contrast,
#> # ⁴lower_95
#>
#> $models
#> # A tibble: 2 × 6
#> Group Variable control model_file model status
#> <chr> <chr> <chr> <chr> <list> <lgl>
#> 1 1 ph.ecog age /var/folders/bj/nw1w4g1j37ddpgb6zmh3sfh… <coxph> TRUE
#> 2 2 ph.ecog age /var/folders/bj/nw1w4g1j37ddpgb6zmh3sfh… <coxph> TRUE
#>
#> attr(,"class")
#> [1] "ezcox"
#>
#> $plot
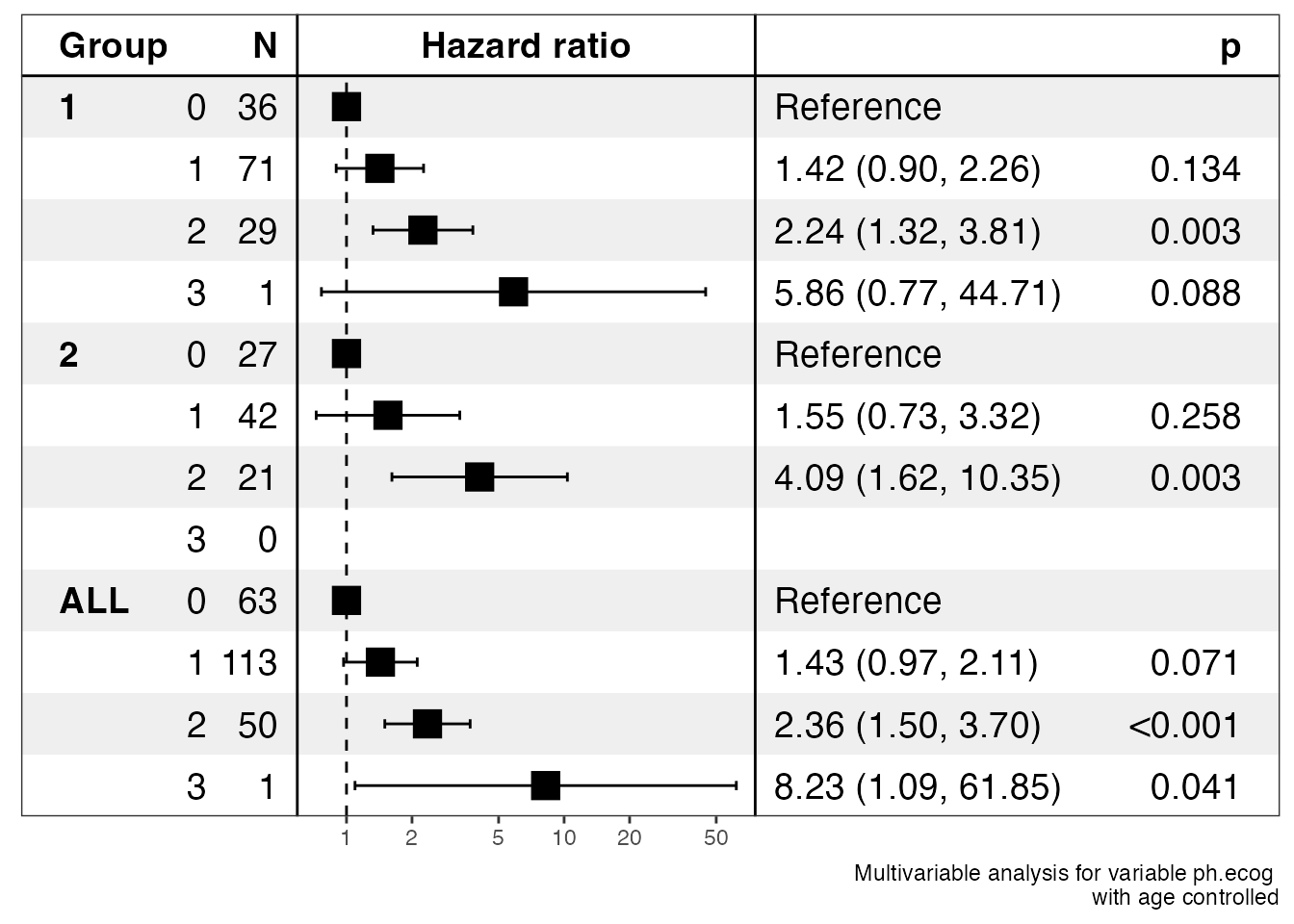
ezcox_group(lung, grp_var = "sex", covariate = "ph.ecog", controls = "age", add_all = TRUE)
#> => Processing variable 1
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable 2
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ALL
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> covariates=NULL but drop_controls=TRUE, detecting controls...
#> Yes. Setting variables to keep...
#> Done.
#> Returns a list containing data and ggplot.
#> $data
#> $stats
#> # A tibble: 12 × 13
#> Group Variable is_cont…¹ contr…² ref_l…³ n_con…⁴ n_ref beta HR lower…⁵
#> <chr> <chr> <lgl> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 ph.ecog FALSE 1 0 71 36 0.353 1.42 0.897
#> 2 1 ph.ecog FALSE 2 0 29 36 0.809 2.24 1.32
#> 3 1 ph.ecog FALSE 3 0 1 36 1.77 5.86 0.767
#> 4 1 ph.ecog TRUE age age 138 138 0.0182 1.02 0.996
#> 5 2 ph.ecog FALSE 1 0 42 27 0.439 1.55 0.726
#> 6 2 ph.ecog FALSE 2 0 21 27 1.41 4.09 1.62
#> 7 2 ph.ecog FALSE 3 0 0 27 NA NA NA
#> 8 2 ph.ecog TRUE age age 90 90 -0.0153 0.985 0.952
#> 9 ALL ph.ecog FALSE 1 0 113 63 0.359 1.43 0.969
#> 10 ALL ph.ecog FALSE 2 0 50 63 0.857 2.36 1.5
#> 11 ALL ph.ecog FALSE 3 0 1 63 2.11 8.23 1.09
#> 12 ALL ph.ecog TRUE age age 228 228 0.0108 1.01 0.992
#> # … with 3 more variables: upper_95 <dbl>, p.value <dbl>, global.pval <dbl>,
#> # and abbreviated variable names ¹is_control, ²contrast_level, ³ref_level,
#> # ⁴n_contrast, ⁵lower_95
#>
#> $models
#> # A tibble: 3 × 6
#> Group Variable control model_file model status
#> <chr> <chr> <chr> <chr> <list> <lgl>
#> 1 1 ph.ecog age /var/folders/bj/nw1w4g1j37ddpgb6zmh3sfh… <coxph> TRUE
#> 2 2 ph.ecog age /var/folders/bj/nw1w4g1j37ddpgb6zmh3sfh… <coxph> TRUE
#> 3 ALL ph.ecog age /var/folders/bj/nw1w4g1j37ddpgb6zmh3sfh… <coxph> TRUE
#>
#> attr(,"class")
#> [1] "ezcox"
#>
#> $plot