Chapter 4 Signature Fit: Sample Signature Exposure Quantification and Analysis
Besides de novo signature discovery shown in previous chapters, another common task is that
you have gotten some reference signatures (either from known database like COSMIC or de novo discovery step), you want to know how these signatures contribute (fit) in a sample. That’s the target of sig_fit().
sig_fit() uses multiple methods to compute exposure of pre-defined signatures from the spectrum of a (can be more) sample. Use ?sig_fit see more detail.
To show how this function works, we use a sample with maximum mutation counts as example data.
i <- which.max(apply(mt_tally$nmf_matrix, 1, sum))
example_mat <- mt_tally$nmf_matrix[i, , drop = FALSE] %>% t()head(example_mat)
#> TCGA-A8-A09G-01A-21W-A019-09
#> A[T>C]A 1
#> C[T>C]A 0
#> G[T>C]A 1
#> T[T>C]A 1
#> A[C>T]A 5
#> C[C>T]A 34.1 Fit Signatures from reference databases
For SBS signatures, users may want to directly use reference signatures from COSMIC database.
sig_fit(example_mat, sig_index = 1:30)
#> ℹ [2021-05-18 23:16:55]: Started.
#> ✓ [2021-05-18 23:16:55]: Signature index detected.
#> ℹ [2021-05-18 23:16:55]: Checking signature database in package.
#> ℹ [2021-05-18 23:16:55]: Checking signature index.
#> ℹ [2021-05-18 23:16:55]: Valid index for db 'legacy':
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30
#> ✓ [2021-05-18 23:16:55]: Database and index checked.
#> ✓ [2021-05-18 23:16:55]: Signature normalized.
#> ℹ [2021-05-18 23:16:55]: Checking row number for catalog matrix and signature matrix.
#> ✓ [2021-05-18 23:16:55]: Checked.
#> ℹ [2021-05-18 23:16:55]: Checking rownames for catalog matrix and signature matrix.
#> ℹ [2021-05-18 23:16:55]: Matrix V and W don't have same orders. Try reordering...
#> ✓ [2021-05-18 23:16:55]: Checked.
#> ✓ [2021-05-18 23:16:55]: Method 'QP' detected.
#> ✓ [2021-05-18 23:16:55]: Corresponding function generated.
#> ℹ [2021-05-18 23:16:55]: Calling function.
#> ℹ [2021-05-18 23:16:55]: Fitting sample: TCGA-A8-A09G-01A-21W-A019-09
#> ✓ [2021-05-18 23:16:55]: Done.
#> ℹ [2021-05-18 23:16:55]: Generating output signature exposures.
#> ✓ [2021-05-18 23:16:55]: Done.
#> ℹ [2021-05-18 23:16:55]: 0.053 secs elapsed.
#> TCGA-A8-A09G-01A-21W-A019-09
#> COSMIC_1 24.215933
#> COSMIC_2 127.164108
#> COSMIC_3 0.000000
#> COSMIC_4 0.000000
#> COSMIC_5 0.000000
#> COSMIC_6 0.000000
#> COSMIC_7 4.907674
#> COSMIC_8 0.000000
#> COSMIC_9 0.000000
#> COSMIC_10 3.584276
#> COSMIC_11 0.000000
#> COSMIC_12 11.062526
#> COSMIC_13 168.298139
#> COSMIC_14 0.000000
#> COSMIC_15 0.000000
#> COSMIC_16 0.000000
#> COSMIC_17 5.578495
#> COSMIC_18 0.000000
#> COSMIC_19 0.000000
#> COSMIC_20 0.000000
#> COSMIC_21 0.000000
#> COSMIC_22 0.000000
#> COSMIC_23 0.000000
#> COSMIC_24 12.084656
#> COSMIC_25 0.000000
#> COSMIC_26 0.000000
#> COSMIC_27 0.000000
#> COSMIC_28 0.000000
#> COSMIC_29 0.000000
#> COSMIC_30 0.104192At default, COSMIC v2 signature database with 30 reference signatures is used (i.e.
sig_db = "legacy"). Setsig_db = "SBS"for COSMIC v3 signature database.
That’s it!
You can set type = "relative" for getting relative exposure.
sig_fit(example_mat, sig_index = 1:30, type = "relative")
#> ℹ [2021-05-18 23:16:55]: Started.
#> ✓ [2021-05-18 23:16:55]: Signature index detected.
#> ℹ [2021-05-18 23:16:55]: Checking signature database in package.
#> ℹ [2021-05-18 23:16:55]: Checking signature index.
#> ℹ [2021-05-18 23:16:55]: Valid index for db 'legacy':
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30
#> ✓ [2021-05-18 23:16:55]: Database and index checked.
#> ✓ [2021-05-18 23:16:55]: Signature normalized.
#> ℹ [2021-05-18 23:16:55]: Checking row number for catalog matrix and signature matrix.
#> ✓ [2021-05-18 23:16:55]: Checked.
#> ℹ [2021-05-18 23:16:55]: Checking rownames for catalog matrix and signature matrix.
#> ℹ [2021-05-18 23:16:55]: Matrix V and W don't have same orders. Try reordering...
#> ✓ [2021-05-18 23:16:55]: Checked.
#> ✓ [2021-05-18 23:16:55]: Method 'QP' detected.
#> ✓ [2021-05-18 23:16:55]: Corresponding function generated.
#> ℹ [2021-05-18 23:16:55]: Calling function.
#> ℹ [2021-05-18 23:16:55]: Fitting sample: TCGA-A8-A09G-01A-21W-A019-09
#> ✓ [2021-05-18 23:16:55]: Done.
#> ℹ [2021-05-18 23:16:55]: Generating output signature exposures.
#> ✓ [2021-05-18 23:16:55]: Done.
#> ℹ [2021-05-18 23:16:55]: 0.04 secs elapsed.
#> TCGA-A8-A09G-01A-21W-A019-09
#> COSMIC_1 0.067832
#> COSMIC_2 0.356202
#> COSMIC_3 0.000000
#> COSMIC_4 0.000000
#> COSMIC_5 0.000000
#> COSMIC_6 0.000000
#> COSMIC_7 0.013747
#> COSMIC_8 0.000000
#> COSMIC_9 0.000000
#> COSMIC_10 0.010040
#> COSMIC_11 0.000000
#> COSMIC_12 0.030987
#> COSMIC_13 0.471423
#> COSMIC_14 0.000000
#> COSMIC_15 0.000000
#> COSMIC_16 0.000000
#> COSMIC_17 0.015626
#> COSMIC_18 0.000000
#> COSMIC_19 0.000000
#> COSMIC_20 0.000000
#> COSMIC_21 0.000000
#> COSMIC_22 0.000000
#> COSMIC_23 0.000000
#> COSMIC_24 0.033851
#> COSMIC_25 0.000000
#> COSMIC_26 0.000000
#> COSMIC_27 0.000000
#> COSMIC_28 0.000000
#> COSMIC_29 0.000000
#> COSMIC_30 0.000292For multiple samples, you can return a data.table, it can be easier to integrate with other information in R.
sig_fit(t(mt_tally$nmf_matrix[1:5, ]), sig_index = 1:30, return_class = "data.table", rel_threshold = 0.05)
#> ℹ [2021-05-18 23:16:55]: Started.
#> ✓ [2021-05-18 23:16:55]: Signature index detected.
#> ℹ [2021-05-18 23:16:55]: Checking signature database in package.
#> ℹ [2021-05-18 23:16:55]: Checking signature index.
#> ℹ [2021-05-18 23:16:55]: Valid index for db 'legacy':
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30
#> ✓ [2021-05-18 23:16:55]: Database and index checked.
#> ✓ [2021-05-18 23:16:55]: Signature normalized.
#> ℹ [2021-05-18 23:16:55]: Checking row number for catalog matrix and signature matrix.
#> ✓ [2021-05-18 23:16:55]: Checked.
#> ℹ [2021-05-18 23:16:55]: Checking rownames for catalog matrix and signature matrix.
#> ℹ [2021-05-18 23:16:55]: Matrix V and W don't have same orders. Try reordering...
#> ✓ [2021-05-18 23:16:55]: Checked.
#> ✓ [2021-05-18 23:16:55]: Method 'QP' detected.
#> ✓ [2021-05-18 23:16:55]: Corresponding function generated.
#> ℹ [2021-05-18 23:16:55]: Calling function.
#> ℹ [2021-05-18 23:16:55]: Fitting sample: TCGA-A1-A0SH-01A-11D-A099-09
#> ℹ [2021-05-18 23:16:55]: Fitting sample: TCGA-A2-A04N-01A-11D-A10Y-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A2-A0CP-01A-11W-A050-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A2-A0EP-01A-52D-A22X-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A2-A0EV-01A-11W-A050-09
#> ✓ [2021-05-18 23:16:56]: Done.
#> ℹ [2021-05-18 23:16:56]: Generating output signature exposures.
#> ✓ [2021-05-18 23:16:56]: Done.
#> ℹ [2021-05-18 23:16:56]: 0.061 secs elapsed.
#> sample COSMIC_1 COSMIC_2 COSMIC_3 COSMIC_4 COSMIC_5 COSMIC_6
#> 1: TCGA-A1-A0SH-01A-11D-A099-09 0.000000 37.420603 13.78689 0.000000 0 12.93472
#> 2: TCGA-A2-A04N-01A-11D-A10Y-09 20.039543 2.888675 0.00000 0.000000 0 0.00000
#> 3: TCGA-A2-A0CP-01A-11W-A050-09 3.648658 0.000000 0.00000 7.083113 0 0.00000
#> 4: TCGA-A2-A0EP-01A-52D-A22X-09 0.000000 0.000000 0.00000 2.492218 0 0.00000
#> 5: TCGA-A2-A0EV-01A-11W-A050-09 6.458422 0.000000 14.83102 0.000000 0 14.78142
#> COSMIC_7 COSMIC_8 COSMIC_9 COSMIC_10 COSMIC_11 COSMIC_12 COSMIC_13 COSMIC_14 COSMIC_15
#> 1: 21.332013 0.00000 0 0.000000 0 0.000000 31.306430 0.000000 0.00000
#> 2: 6.865345 12.11501 0 0.000000 0 0.000000 0.000000 0.000000 0.00000
#> 3: 10.348536 0.00000 0 0.000000 0 0.000000 0.000000 0.000000 18.37734
#> 4: 2.156319 0.00000 0 0.000000 0 1.334731 4.654227 6.728415 0.00000
#> 5: 21.963952 0.00000 0 7.978962 0 0.000000 5.713563 0.000000 0.00000
#> COSMIC_16 COSMIC_17 COSMIC_18 COSMIC_19 COSMIC_20 COSMIC_21 COSMIC_22 COSMIC_23 COSMIC_24
#> 1: 0 0 12.007682 0 0.000000 0.000000 0 0.00000 0
#> 2: 0 0 0.000000 0 7.516444 0.000000 0 0.00000 0
#> 3: 0 0 4.384106 0 0.000000 0.000000 0 0.00000 0
#> 4: 0 0 0.000000 0 0.000000 0.000000 0 1.26778 0
#> 5: 0 0 0.000000 0 0.000000 4.311951 0 0.00000 0
#> COSMIC_25 COSMIC_26 COSMIC_27 COSMIC_28 COSMIC_29 COSMIC_30
#> 1: 0 0 0 0 0.000000 0
#> 2: 0 0 0 0 0.000000 0
#> 3: 0 0 0 0 4.776321 0
#> 4: 0 0 0 0 0.000000 0
#> 5: 0 0 0 0 0.000000 0When you set multiple signatures, we recommend setting rel_threshold option, which will set exposure of a signature to 0 if its relative exposure in a sample less than the rel_threshold.
4.2 Fit Custom Signatures
We have already determined the SBS signatures before. Here we can set them to sig option.
sig_fit(example_mat, sig = mt_sig2)
#> ℹ [2021-05-18 23:16:56]: Started.
#> ℹ [2021-05-18 23:16:56]: Signature index not detected.
#> ✓ [2021-05-18 23:16:56]: Signature object detected.
#> ✓ [2021-05-18 23:16:56]: Database and index checked.
#> ✓ [2021-05-18 23:16:56]: Signature normalized.
#> ℹ [2021-05-18 23:16:56]: Checking row number for catalog matrix and signature matrix.
#> ✓ [2021-05-18 23:16:56]: Checked.
#> ℹ [2021-05-18 23:16:56]: Checking rownames for catalog matrix and signature matrix.
#> ✓ [2021-05-18 23:16:56]: Checked.
#> ✓ [2021-05-18 23:16:56]: Method 'QP' detected.
#> ✓ [2021-05-18 23:16:56]: Corresponding function generated.
#> ℹ [2021-05-18 23:16:56]: Calling function.
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A8-A09G-01A-21W-A019-09
#> ✓ [2021-05-18 23:16:56]: Done.
#> ℹ [2021-05-18 23:16:56]: Generating output signature exposures.
#> ✓ [2021-05-18 23:16:56]: Done.
#> ℹ [2021-05-18 23:16:56]: 0.032 secs elapsed.
#> TCGA-A8-A09G-01A-21W-A019-09
#> Sig1 0
#> Sig2 0
#> Sig3 3574.3 Performance Comparison
Now that we can use sig_fit for getting optimal exposures, we can compare the RSS between raw matrix and the reconstructed matrix either by NMF and sig_fit().
i.e.
\[ RSS = \sum(\hat H - H)^2 \]
## Exposure got from NMF
sum((apply(mt_sig2$Signature, 2, function(x) x / sum(x)) %*% mt_sig2$Exposure - t(mt_tally$nmf_matrix))^2)
#> [1] 8891.978## Exposure optimized by sig_fit
H_estimate <- apply(mt_sig2$Signature, 2, function(x) x / sum(x)) %*% sig_fit(t(mt_tally$nmf_matrix), sig = mt_sig2)
#> ℹ [2021-05-18 23:16:56]: Started.
#> ℹ [2021-05-18 23:16:56]: Signature index not detected.
#> ✓ [2021-05-18 23:16:56]: Signature object detected.
#> ✓ [2021-05-18 23:16:56]: Database and index checked.
#> ✓ [2021-05-18 23:16:56]: Signature normalized.
#> ℹ [2021-05-18 23:16:56]: Checking row number for catalog matrix and signature matrix.
#> ✓ [2021-05-18 23:16:56]: Checked.
#> ℹ [2021-05-18 23:16:56]: Checking rownames for catalog matrix and signature matrix.
#> ✓ [2021-05-18 23:16:56]: Checked.
#> ✓ [2021-05-18 23:16:56]: Method 'QP' detected.
#> ✓ [2021-05-18 23:16:56]: Corresponding function generated.
#> ℹ [2021-05-18 23:16:56]: Calling function.
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A1-A0SH-01A-11D-A099-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A2-A04N-01A-11D-A10Y-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A2-A0CP-01A-11W-A050-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A2-A0EP-01A-52D-A22X-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A2-A0EV-01A-11W-A050-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A2-A0SX-01A-12D-A099-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A2-A0T7-01A-21D-A099-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A2-A0YF-01A-21D-A10G-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A2-A25F-01A-11D-A167-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A2-A3XW-01A-11D-A23C-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A2-A4S1-01A-21D-A25Q-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A7-A0D9-01A-31W-A071-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A7-A13F-01A-11D-A12Q-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A7-A5ZV-01A-11D-A28B-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A8-A06P-01A-11W-A019-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A8-A076-01A-21W-A019-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A8-A07W-01A-11W-A019-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A8-A084-01A-21W-A019-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A8-A08S-01A-11W-A050-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A8-A09G-01A-21W-A019-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A8-A0A4-01A-11W-A019-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A8-A0AB-01A-11W-A050-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AC-A2B8-01A-11D-A17D-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AC-A2FO-01A-11D-A17W-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AC-A3YI-01A-21D-A23C-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AC-A8OS-01A-12D-A41F-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AN-A0FK-01A-11W-A050-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AN-A0FT-01A-11W-A050-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AN-A0XO-01A-11D-A10G-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AO-A1KS-01A-11D-A13L-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AQ-A54O-01A-11D-A25Q-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AQ-A7U7-01A-22D-A351-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AR-A0TP-01A-11D-A099-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AR-A0U3-01A-11D-A10G-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AR-A1AH-01A-11D-A12B-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AR-A1AJ-01A-21D-A12Q-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AR-A1AN-01A-11D-A12Q-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AR-A24N-01A-11D-A167-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AR-A252-01A-11D-A167-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AR-A2LL-01A-11D-A17W-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-AR-A2LO-01A-31D-A18P-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-B6-A0IE-01A-11W-A050-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-B6-A0IM-01A-11W-A050-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-B6-A0IP-01A-11D-A045-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-B6-A0RV-01A-11D-A099-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-B6-A0WZ-01A-11D-A10G-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-B6-A0X1-01A-11D-A10G-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-B6-A1KC-01B-11D-A159-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-B6-A401-01A-11D-A23C-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-B6-A40C-01A-11D-A23C-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-BH-A0AV-01A-31D-A10Y-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-BH-A0BT-01A-11D-A12Q-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-BH-A0DL-01A-11D-A10Y-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-BH-A0DO-01B-11D-A12B-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-BH-A0DT-01A-21D-A12B-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-BH-A0GY-01A-11W-A071-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-BH-A0H6-01A-21W-A071-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-BH-A18K-01A-11D-A12B-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-BH-A1FU-01A-11D-A14G-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-BH-A202-01A-11D-A14K-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-BH-A5IZ-01A-11D-A27P-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-BH-A6R8-01A-21D-A33E-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-BH-A8G0-01A-11D-A351-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-C8-A131-01A-11D-A10Y-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-D8-A147-01A-11D-A10Y-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-D8-A1JG-01B-11D-A13L-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-D8-A1JH-01A-11D-A188-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-D8-A1JJ-01A-31D-A14K-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-D8-A1JT-01A-31D-A13L-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-D8-A1JU-01A-11D-A13L-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-D8-A1X7-01A-11D-A14K-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-D8-A1X8-01A-11D-A14K-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-D8-A1XL-01A-11D-A14K-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-D8-A27V-01A-12D-A17D-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E2-A108-01A-13D-A10M-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E2-A10F-01A-11D-A10M-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E2-A14T-01A-11D-A10Y-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E2-A152-01A-11D-A12B-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E2-A15D-01A-11D-A10Y-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E2-A15L-01A-11D-A12B-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E2-A1BD-01A-11D-A12Q-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E2-A1IH-01A-11D-A188-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E2-A1II-01A-11D-A142-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E2-A1IJ-01A-11D-A142-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E2-A1L6-01A-11D-A13L-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E2-A9RU-01A-11D-A41F-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E9-A1NE-01A-21D-A14K-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E9-A22A-01A-11D-A159-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E9-A22E-01A-11D-A159-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E9-A3QA-01A-61D-A228-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-E9-A5FL-01A-11D-A27P-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-EW-A1PA-01A-11D-A142-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-EW-A1PH-01A-11D-A14K-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-GM-A2DB-01A-31D-A19Y-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-LD-A9QF-01A-32D-A41F-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-LL-A5YP-01A-21D-A28B-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-LL-A73Z-01A-11D-A32I-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-OL-A5RY-01A-21D-A28B-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-PE-A5DD-01A-12D-A27P-09
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-S3-AA17-01A-11D-A41F-09
#> ✓ [2021-05-18 23:16:56]: Done.
#> ℹ [2021-05-18 23:16:56]: Generating output signature exposures.
#> ✓ [2021-05-18 23:16:56]: Done.
#> ℹ [2021-05-18 23:16:56]: 0.243 secs elapsed.
H_estimate <- apply(H_estimate, 2, function(x) ifelse(is.nan(x), 0, x))
H_real <- t(mt_tally$nmf_matrix)
sum((H_estimate - H_real)^2)
#> [1] 8242.634.4 Estimate Exposure Stability by Bootstrap
This feature is based on sig_fit(), it uses the resampling data of original input and runs sig_fit() multiple times to estimate the exposure. Bootstrap replicates >= 100 is recommended, here I just use 10 times for illustration.
bt_result <- sig_fit_bootstrap_batch(example_mat, sig = mt_sig2, n = 10)
#> ℹ [2021-05-18 23:16:56]: Batch Bootstrap Signature Exposure Analysis Started.
#> ℹ [2021-05-18 23:16:56]: Samples to be filtered out:
#> ℹ [2021-05-18 23:16:56]: Finding optimal exposures (&errors) for different methods.
#> ℹ [2021-05-18 23:16:56]: Calling method `QP`.
#> ℹ [2021-05-18 23:16:56]: Started.
#> ℹ [2021-05-18 23:16:56]: Signature index not detected.
#> ✓ [2021-05-18 23:16:56]: Signature object detected.
#> ✓ [2021-05-18 23:16:56]: Database and index checked.
#> ✓ [2021-05-18 23:16:56]: Signature normalized.
#> ℹ [2021-05-18 23:16:56]: Checking row number for catalog matrix and signature matrix.
#> ✓ [2021-05-18 23:16:56]: Checked.
#> ℹ [2021-05-18 23:16:56]: Checking rownames for catalog matrix and signature matrix.
#> ✓ [2021-05-18 23:16:56]: Checked.
#> ✓ [2021-05-18 23:16:56]: Method 'QP' detected.
#> ✓ [2021-05-18 23:16:56]: Corresponding function generated.
#> ℹ [2021-05-18 23:16:56]: Calling function.
#> ℹ [2021-05-18 23:16:56]: Fitting sample: TCGA-A8-A09G-01A-21W-A019-09
#> ✓ [2021-05-18 23:16:56]: Done.
#> ℹ [2021-05-18 23:16:56]: Generating output signature exposures.
#> ✓ [2021-05-18 23:16:56]: Done.
#> ℹ [2021-05-18 23:16:57]: Calculating errors (Frobenius Norm).
#> ✓ [2021-05-18 23:16:57]: Done.
#> ℹ [2021-05-18 23:16:57]: 0.072 secs elapsed.
#> ℹ [2021-05-18 23:16:57]: Getting bootstrap exposures (&errors/similarity) for different methods.
#> ℹ [2021-05-18 23:16:57]: This step is time consuming, please be patient.
#> ℹ [2021-05-18 23:16:57]: Processing sample `TCGA-A8-A09G-01A-21W-A019-09`.
#> ℹ [2021-05-18 23:16:57]: Started.
#> ℹ [2021-05-18 23:16:57]: Checking catalog.
#> ✓ [2021-05-18 23:16:57]: Done.
#> ℹ [2021-05-18 23:16:57]: About to start bootstrap.
#>
→ Bootstrapping 10 times.
→ Total 10 times, starting no.1.
→ Total 10 times, starting no.2.
→ Total 10 times, starting no.3.
→ Total 10 times, starting no.4.
→ Total 10 times, starting no.5.
→ Total 10 times, starting no.6.
→ Total 10 times, starting no.7.
→ Total 10 times, starting no.8.
→ Total 10 times, starting no.9.
→ Total 10 times, starting no.10.
Bootstrap done.
#> ✓ [2021-05-18 23:16:57]: Signature exposures collected.
#> ✓ [2021-05-18 23:16:57]: Errors and similarity collected.
#> ✓ [2021-05-18 23:16:57]: Done.
#> ℹ [2021-05-18 23:16:57]: 0.69 secs elapsed.
#> ✓ [2021-05-18 23:16:58]: Gotten.
#> ℹ [2021-05-18 23:16:58]: Reporting p values...
#> ℹ [2021-05-18 23:16:58]: Started.
#> ✓ [2021-05-18 23:16:58]: Batch mode enabled.
#> ✓ [2021-05-18 23:16:58]: Done.
#> ℹ [2021-05-18 23:16:58]: 0.013 secs elapsed.
#> ✓ [2021-05-18 23:16:58]: Done.
#> ℹ [2021-05-18 23:16:58]: Cleaning results...
#> ✓ [2021-05-18 23:16:58]: Outputing.
#> ℹ [2021-05-18 23:16:58]: Total 1.454 secs elapsed.
bt_result
#> $expo
#> method sample sig exposure type
#> 1: QP TCGA-A8-A09G-01A-21W-A019-09 Sig1 0.000000 optimal
#> 2: QP TCGA-A8-A09G-01A-21W-A019-09 Sig2 0.000000 optimal
#> 3: QP TCGA-A8-A09G-01A-21W-A019-09 Sig3 357.000000 optimal
#> 4: QP TCGA-A8-A09G-01A-21W-A019-09 Sig1 1.998810 Rep_1
#> 5: QP TCGA-A8-A09G-01A-21W-A019-09 Sig2 0.000000 Rep_1
#> 6: QP TCGA-A8-A09G-01A-21W-A019-09 Sig3 355.001190 Rep_1
#> 7: QP TCGA-A8-A09G-01A-21W-A019-09 Sig1 0.000000 Rep_2
#> 8: QP TCGA-A8-A09G-01A-21W-A019-09 Sig2 0.324300 Rep_2
#> 9: QP TCGA-A8-A09G-01A-21W-A019-09 Sig3 356.675700 Rep_2
#> 10: QP TCGA-A8-A09G-01A-21W-A019-09 Sig1 0.000000 Rep_3
#> 11: QP TCGA-A8-A09G-01A-21W-A019-09 Sig2 3.656600 Rep_3
#> 12: QP TCGA-A8-A09G-01A-21W-A019-09 Sig3 353.343400 Rep_3
#> 13: QP TCGA-A8-A09G-01A-21W-A019-09 Sig1 0.000000 Rep_4
#> 14: QP TCGA-A8-A09G-01A-21W-A019-09 Sig2 20.198785 Rep_4
#> 15: QP TCGA-A8-A09G-01A-21W-A019-09 Sig3 336.801215 Rep_4
#> 16: QP TCGA-A8-A09G-01A-21W-A019-09 Sig1 0.000000 Rep_5
#> 17: QP TCGA-A8-A09G-01A-21W-A019-09 Sig2 0.000000 Rep_5
#> 18: QP TCGA-A8-A09G-01A-21W-A019-09 Sig3 357.000000 Rep_5
#> 19: QP TCGA-A8-A09G-01A-21W-A019-09 Sig1 0.000000 Rep_6
#> 20: QP TCGA-A8-A09G-01A-21W-A019-09 Sig2 19.971588 Rep_6
#> 21: QP TCGA-A8-A09G-01A-21W-A019-09 Sig3 337.028412 Rep_6
#> 22: QP TCGA-A8-A09G-01A-21W-A019-09 Sig1 0.000000 Rep_7
#> 23: QP TCGA-A8-A09G-01A-21W-A019-09 Sig2 0.111127 Rep_7
#> 24: QP TCGA-A8-A09G-01A-21W-A019-09 Sig3 356.888873 Rep_7
#> 25: QP TCGA-A8-A09G-01A-21W-A019-09 Sig1 0.000000 Rep_8
#> 26: QP TCGA-A8-A09G-01A-21W-A019-09 Sig2 1.656231 Rep_8
#> 27: QP TCGA-A8-A09G-01A-21W-A019-09 Sig3 355.343769 Rep_8
#> 28: QP TCGA-A8-A09G-01A-21W-A019-09 Sig1 0.000000 Rep_9
#> 29: QP TCGA-A8-A09G-01A-21W-A019-09 Sig2 0.000000 Rep_9
#> 30: QP TCGA-A8-A09G-01A-21W-A019-09 Sig3 357.000000 Rep_9
#> 31: QP TCGA-A8-A09G-01A-21W-A019-09 Sig1 9.636951 Rep_10
#> 32: QP TCGA-A8-A09G-01A-21W-A019-09 Sig2 3.898671 Rep_10
#> 33: QP TCGA-A8-A09G-01A-21W-A019-09 Sig3 343.464378 Rep_10
#> method sample sig exposure type
#>
#> $error
#> method sample errors type
#> 1: QP TCGA-A8-A09G-01A-21W-A019-09 18.549 optimal
#> 2: QP TCGA-A8-A09G-01A-21W-A019-09 18.597 Rep_1
#> 3: QP TCGA-A8-A09G-01A-21W-A019-09 18.555 Rep_2
#> 4: QP TCGA-A8-A09G-01A-21W-A019-09 18.656 Rep_3
#> 5: QP TCGA-A8-A09G-01A-21W-A019-09 20.243 Rep_4
#> 6: QP TCGA-A8-A09G-01A-21W-A019-09 18.549 Rep_5
#> 7: QP TCGA-A8-A09G-01A-21W-A019-09 20.210 Rep_6
#> 8: QP TCGA-A8-A09G-01A-21W-A019-09 18.551 Rep_7
#> 9: QP TCGA-A8-A09G-01A-21W-A019-09 18.586 Rep_8
#> 10: QP TCGA-A8-A09G-01A-21W-A019-09 18.549 Rep_9
#> 11: QP TCGA-A8-A09G-01A-21W-A019-09 19.305 Rep_10
#>
#> $cosine
#> method sample cosine type
#> 1: QP TCGA-A8-A09G-01A-21W-A019-09 0.988326 optimal
#> 2: QP TCGA-A8-A09G-01A-21W-A019-09 0.985484 Rep_1
#> 3: QP TCGA-A8-A09G-01A-21W-A019-09 0.971921 Rep_2
#> 4: QP TCGA-A8-A09G-01A-21W-A019-09 0.980851 Rep_3
#> 5: QP TCGA-A8-A09G-01A-21W-A019-09 0.986036 Rep_4
#> 6: QP TCGA-A8-A09G-01A-21W-A019-09 0.977990 Rep_5
#> 7: QP TCGA-A8-A09G-01A-21W-A019-09 0.979053 Rep_6
#> 8: QP TCGA-A8-A09G-01A-21W-A019-09 0.975565 Rep_7
#> 9: QP TCGA-A8-A09G-01A-21W-A019-09 0.983459 Rep_8
#> 10: QP TCGA-A8-A09G-01A-21W-A019-09 0.982522 Rep_9
#> 11: QP TCGA-A8-A09G-01A-21W-A019-09 0.978299 Rep_10
#>
#> $p_val
#> sample method threshold sig p_value
#> 1: TCGA-A8-A09G-01A-21W-A019-09 QP 0.05 Sig1 1.000000e+00
#> 2: TCGA-A8-A09G-01A-21W-A019-09 QP 0.05 Sig2 9.996431e-01
#> 3: TCGA-A8-A09G-01A-21W-A019-09 QP 0.05 Sig3 3.265198e-16You can plot the result very easily with functions provided by sigminer.
show_sig_bootstrap_exposure(bt_result, sample = "TCGA-A8-A09G-01A-21W-A019-09")
#> ℹ [2021-05-18 23:16:58]: Started.
#> ℹ [2021-05-18 23:16:58]: Plotting.
#> ℹ [2021-05-18 23:16:58]: 0.056 secs elapsed.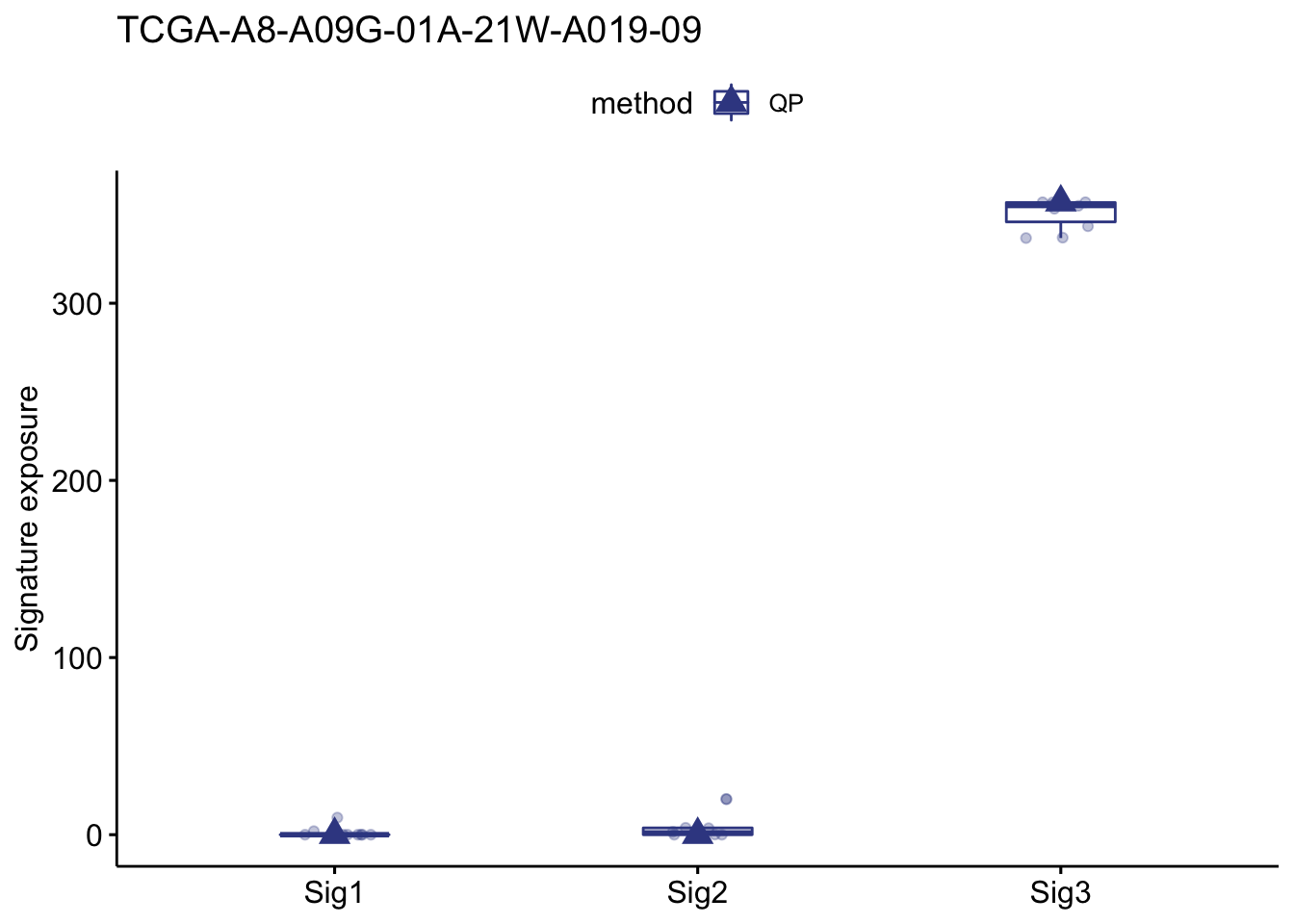
show_sig_bootstrap_error(bt_result, sample = "TCGA-A8-A09G-01A-21W-A019-09")
#> ℹ [2021-05-18 23:16:59]: Started.
#> ℹ [2021-05-18 23:16:59]: Plotting.
#> ℹ [2021-05-18 23:16:59]: 0.032 secs elapsed.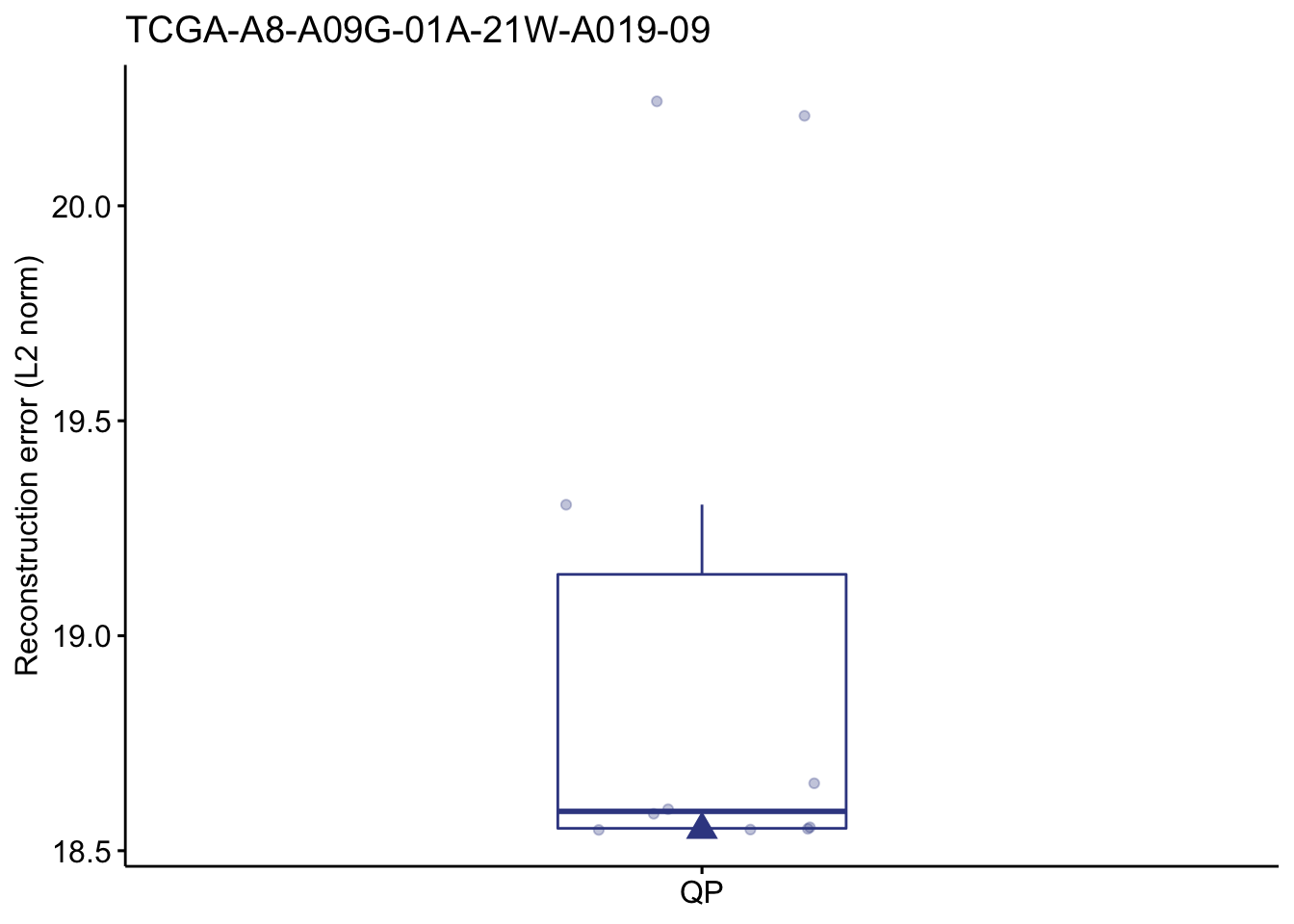
show_sig_bootstrap_stability(bt_result)
#> ℹ [2021-05-18 23:16:59]: Started.
#> ℹ [2021-05-18 23:16:59]: Plotting.
#> ℹ [2021-05-18 23:16:59]: 0.079 secs elapsed.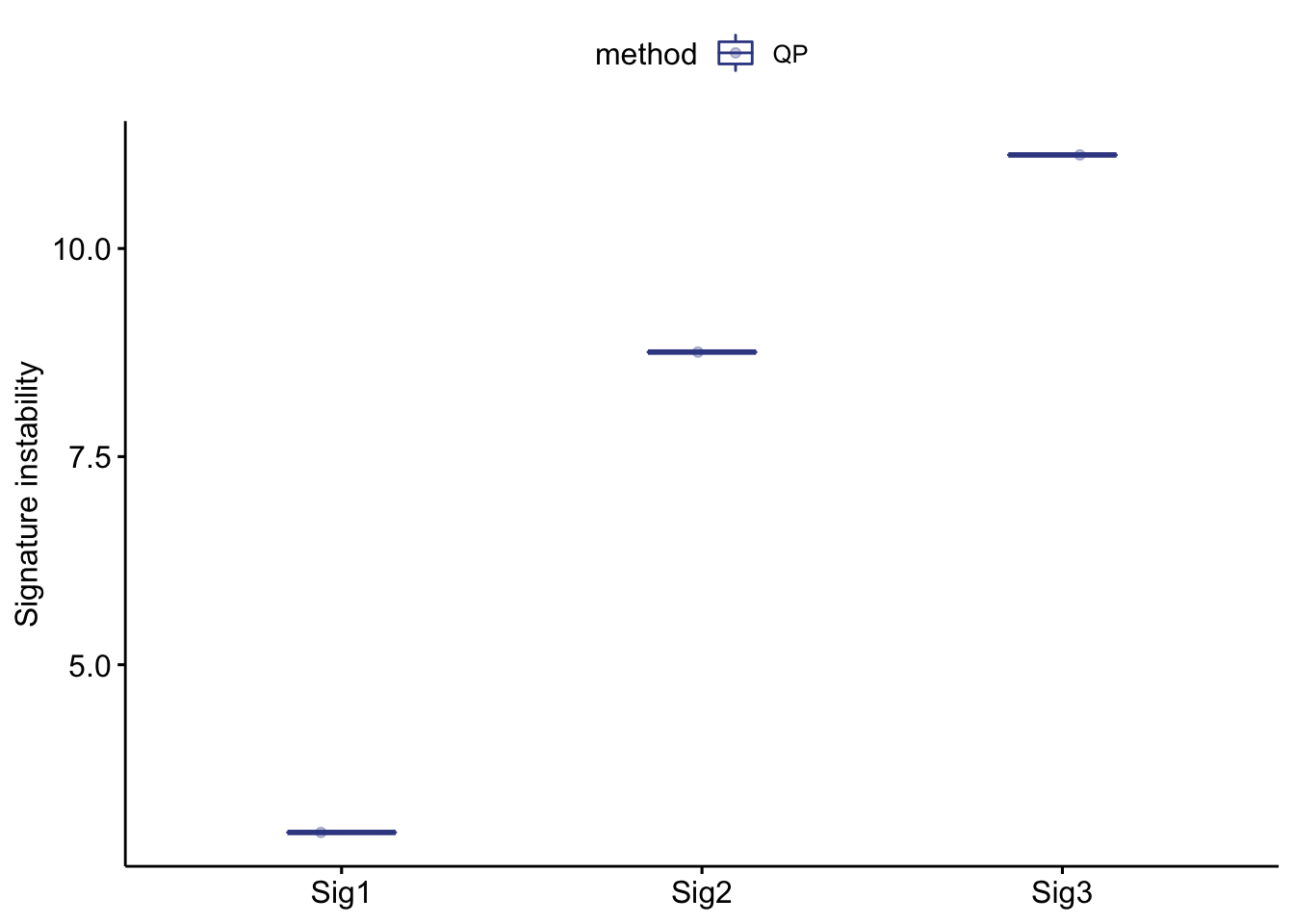 P values have been calculated under specified relative exposure cutoff (0.05 at default).
P values have been calculated under specified relative exposure cutoff (0.05 at default).
The result indicates Sig3 is very stable.